Figure 7.
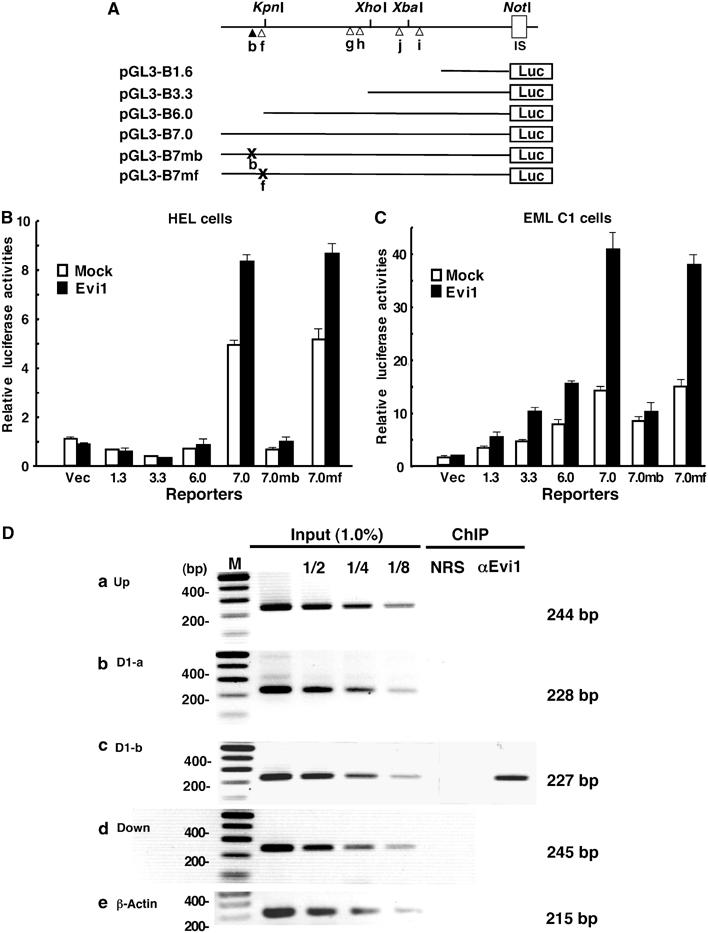
Evi1 directly binds to the promoter region of GATA-2 and thus enhances the GATA-2 transcription. (A) Structure of reporter plasmids of the GATA-2 promoter region. Various lengths (1.6, 3.3, 6.0 and 7.0 kb) of the promoter region and 7.0 kb of the GATA-2 promoter region containing mutations in the binding site (b) (from GACATGGACA to GACATGGAGA) (7mb) or in the binding site (f) (from GATGAG to TATGAG) (7mf) were inserted upstream of the luciferase gene in a promoter-less reporter plasmid (pGL3-Basic). The top line shows six possible binding sites (b, f–h, j and l) and four unique restriction sites (KpnI, XhoI, XbaI and NotI) located upstream of the IS exon. (B) GATA-2 promoter activity and transactivation by exogenous Evi1 in HEL cells. Various GATA-2 IS promoter-LUC reporter constructs (pGL3-B1.6, 3.3, 6.0 and 7.0) were transfected into HEL cells with (black bar) or without (white bar) an Evi1 expression vector. A mock vector (pGL3-B) was used as a control (Vec). (C) GATA-2 promoter activity and transactivation by exogenous Evi1 in EML C1 cells. The experiment was performed as in HEL cells. (D) Specific DNA binding of Evi1 to site b detected by ChIP. Five genomic DNA fragments containing each possible DNA-binding site (D1-a or D1-b) were amplified from the genomic DNA from fixed EML C1 cells after immunoprecipitation with normal rabbit serum (NRS) or with anti-Evi1 antibody (αEvi1). The localization of the amplified regions in GATA-2 promoter is shown in Supplementary Figure 6A. For controls, each genomic region was amplified from purified DNA with the indicated dilutions after formaldehyde fixation and sonication (Input).
