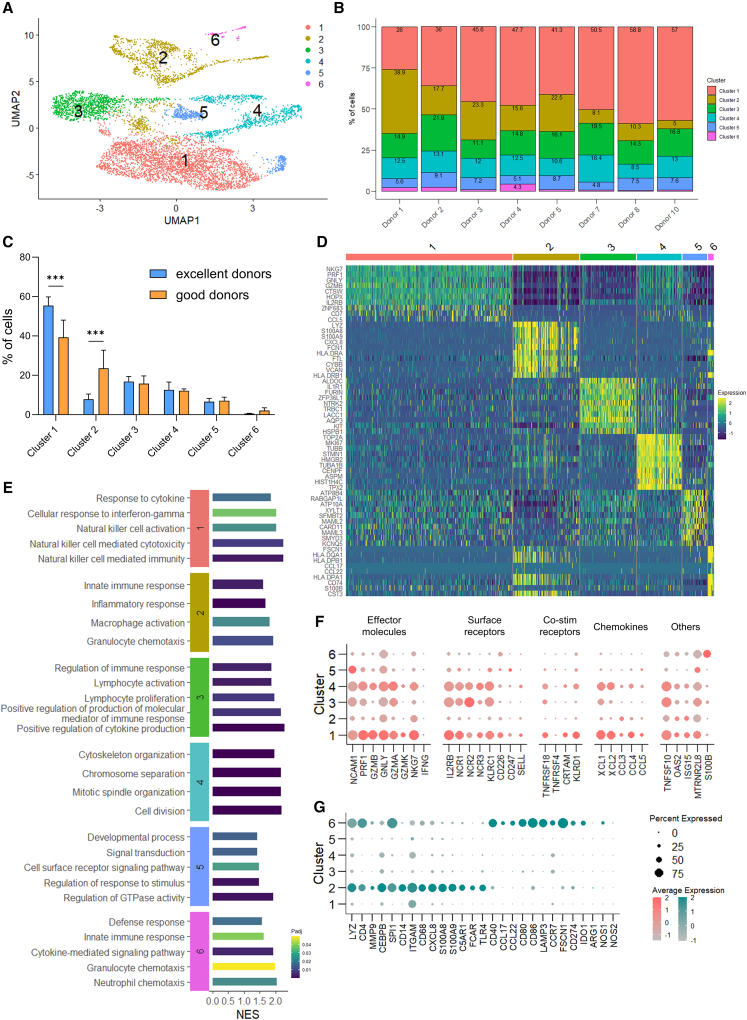
Figure 3.
scRNA-seq reveals early enrichment of NK effector-like cells and lack of myeloid lineage cells during differentiation of excellent donors
(A) UMAP of cluster distribution of cells, analyzed with Leiden algorithm on day 28 from 8 donors. (B) Relative distribution of clusters per donor, expressed as percentage of total. (C) Distribution of clusters across excellent and good donors (n = 3 and 5, respectively), expressed as percentage of total. Clusters are ranked by overall size. Data are shown as mean ± SD. Statistical analysis was performed using two-way ANOVA with Šídák correction for multiple comparisons. Significance is shown as ∗∗∗p <0.001. (D) Heatmap showing the top 10 markers of each cluster identified by differential expression analysis. (E) Selected GO Biological Process pathways enriched in each cluster. (F and G) Dot plots of (F) selected NK cell-related genes and (G) selected myeloid-related genes. The size of the dot is proportional to the percentage of cells expressing the gene, while the color scale indicates the average scaled gene expression. Excellent and good donors are labeled with different colors. UMAP, Uniform Manifold Approximation and Projection; GO, Gene Ontology; NES, normalized enrichment score; Padj, adjusted p value.