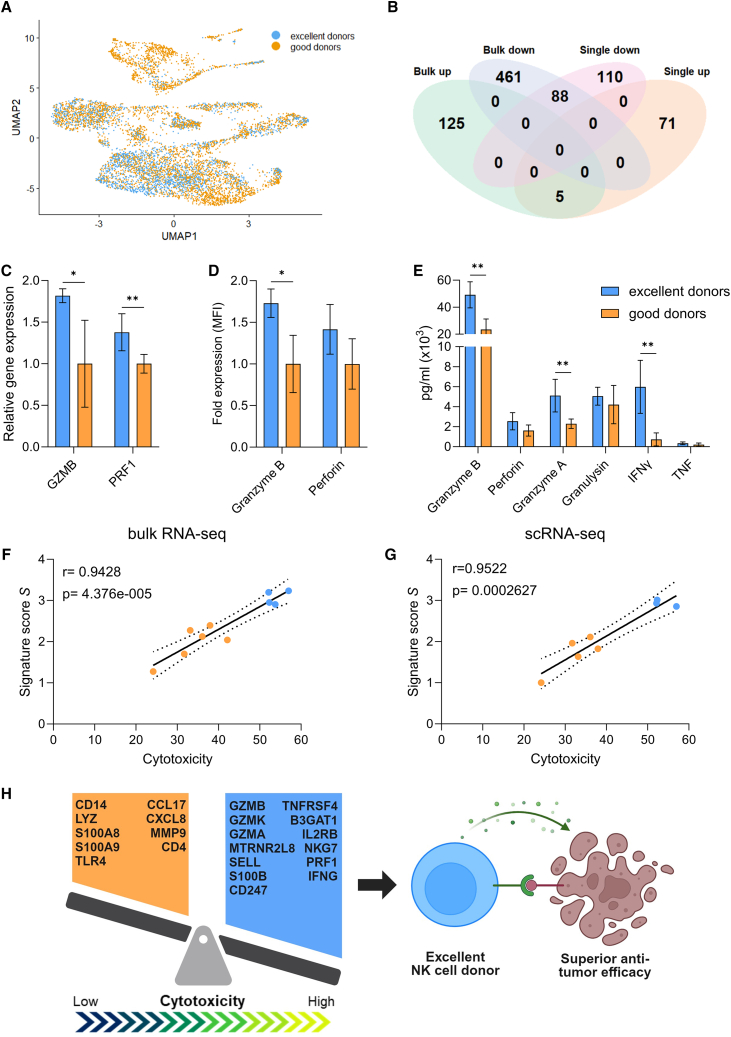
Figure 5.
Identification of a multi-factorial gene expression signature predicting donor cytotoxic potential
(A) UMAP of excellent and good donor distribution. No major separation is distinguishable. (B) Overlap of differentially expressed genes between excellent and good donors identified with bulk and scRNA-seq. (C) Relative mRNA expression of GZMB and PRF1 of day 35 cells, analyzed by qPCR on the 10 donors. The good group is used as control. (D) Intracellular protein levels of granzyme B and perforin of day 35 cells, analyzed via flow cytometry on 9 donors. The good group is used as control. (E) Detection of effector molecules in the supernatant after 5 h co-culture with K562 target cells, analyzed via flow cytometry on 9 donors. In (C)–(E), data are shown as mean ± SD and statistical analysis was performed using unpaired t test, with Holm-Šídák correction for multiple comparisons. Significance is shown as ∗p < 0.05, ∗∗p < 0.01. (F and G) Linear correlation analysis between the mean cytotoxicity value, calculated across 4 target cell lines, and the signature score S, from (F) bulk RNA-seq (10 donors) and (G) scRNA-seq (8 donors). (H) Summary of the main findings from this study. Excellent and good NK cell-related genes, contributing to the predictive signature, are listed in the blue and orange regions, respectively. MFI, mean fluorescence intensity.