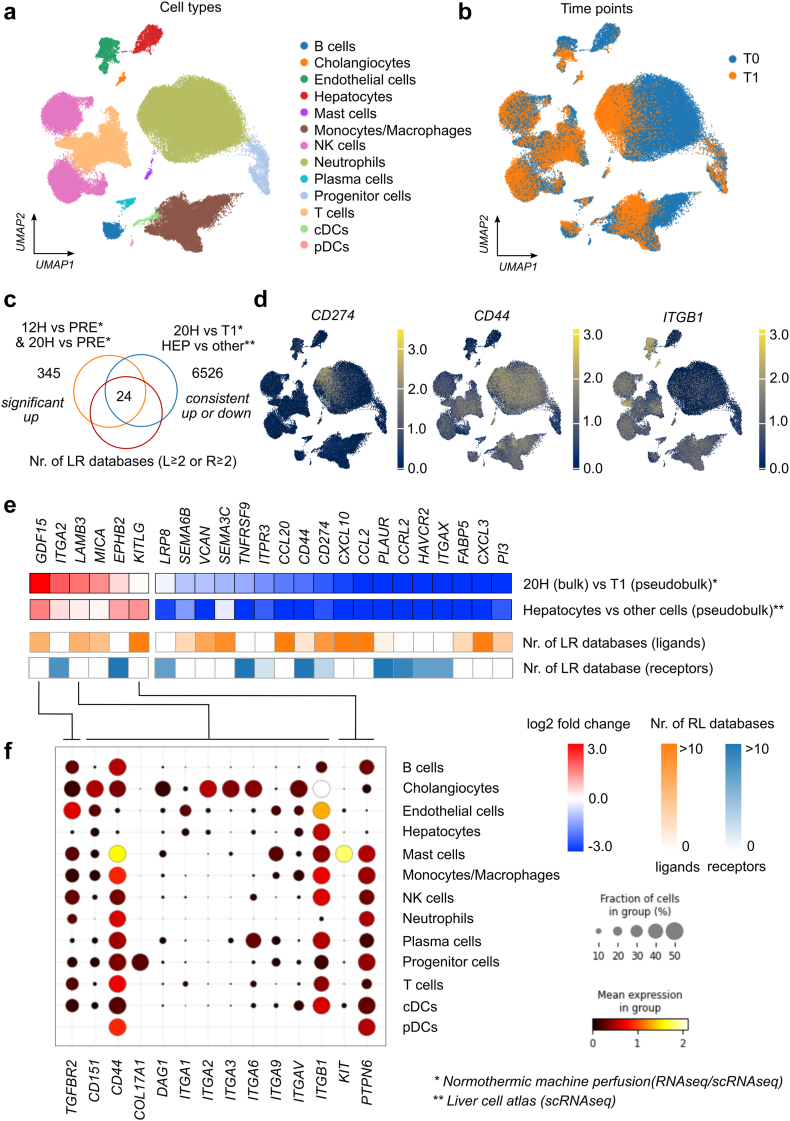
Fig. 3.
Analyses of crosstalk between hepatocytes and other cells during normothermic machine perfusion (NMP) using single-cell RNA sequencing data. (a) UMAP showing clusters of single-cell data with cell type annotation. (b) Cells from livers during NMP (T1 refers to end of NMP and T0 refers to baseline before NMP also named PRE) (c) Venn diagram indicating 24 genes which are consistently up- (log2FC >0) or down regulated (log2FC <0) 1) between bulk RNA sequencing expression data after 20 h NMP with pseudo-bulk data from scRNAseq at T1, 2) between hepatocytes and all other cell types using the pseudo-bulk data from single-cell analyses of normal liver tissue, and 3) are annotated as a ligand or receptor in at least two out of 13 curated ligand-receptor databases. (d) UMAP plot indicating expression of selected genes (receptors). (e) Heatmap of the 24 genes indicating differential expression (log2 fold change) for the two comparisons and number of annotation as ligand or receptor in ligand-receptor databases. (f) Expression of respective receptors in various cell types during liver NMP based on scRNAseq data according to the legend at the right.