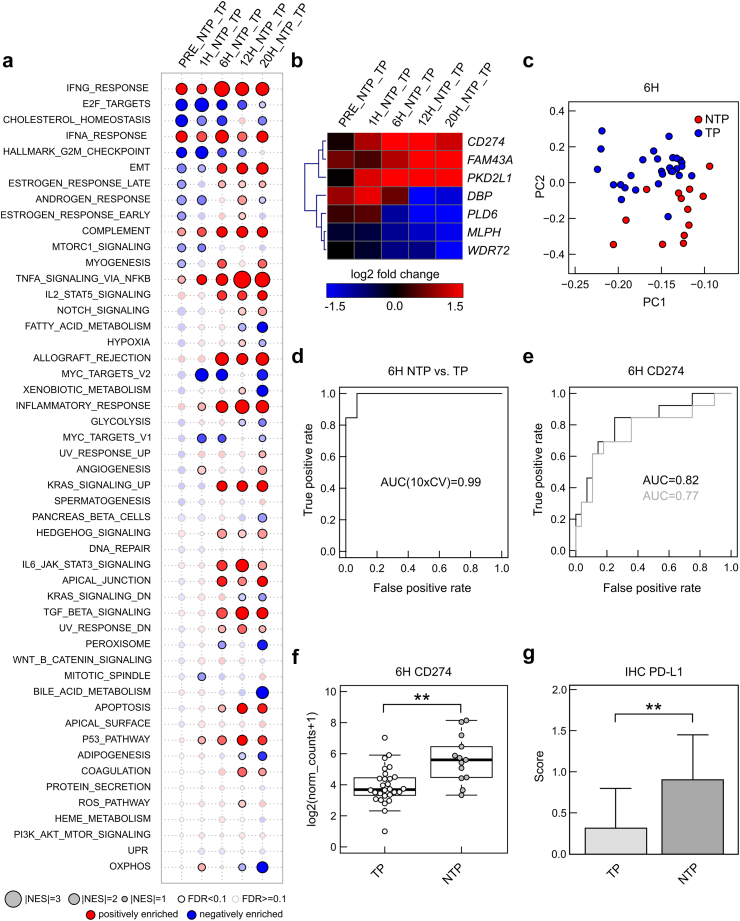
Fig. 6.
Differential genes (signatures) between non-transplanted livers (NTP) and transplanted livers (TP). (a) Gene set enrichment analyses indicating positively (red) or negatively (blue) enrichment of hallmark gene sets in differential expression of genes in NTP vs TP at different time points (size indicate normalized enrichment score, solid border indicates significant enrichment with false discovery rate (FDR) <0.1). (b) Heatmap indicating log2 fold changes of 7 genes as identified using regularized logistic regression (LASSO). (c) Principal component analysis showing separation of NTP vs TP at 6 h. (d) Receiver operating characteristics of this 7-gene model and area under curve (AUC) (mean of 10-fold cross validation). (e) Receiver operating characteristics (ROC) curve and AUC for the CD274 gene (grey line: leave-one-out cross validated logistic regression model). (f) Boxplot indicating differences in gene expression of CD274 (log2 from normalized counts adding pseudocount of 1) between NTP (n = 13) and TP (n = 28) at 6 h. ∗∗ FDR<0.01 (test based on a negative binomial distribution using DESeq2 and correction based on the FDR according to the Benjamini-Hochberg method) (g) PD-L1 protein expression (encoded by CD274 gene) in immune cells using immunohistochemical stains in liver biopsies at 6 h in NTP (n = 13) and TP (n = 28) livers. ∗∗p < 0.05 (two-sided Student's t test, error bars indicate standard deviation).