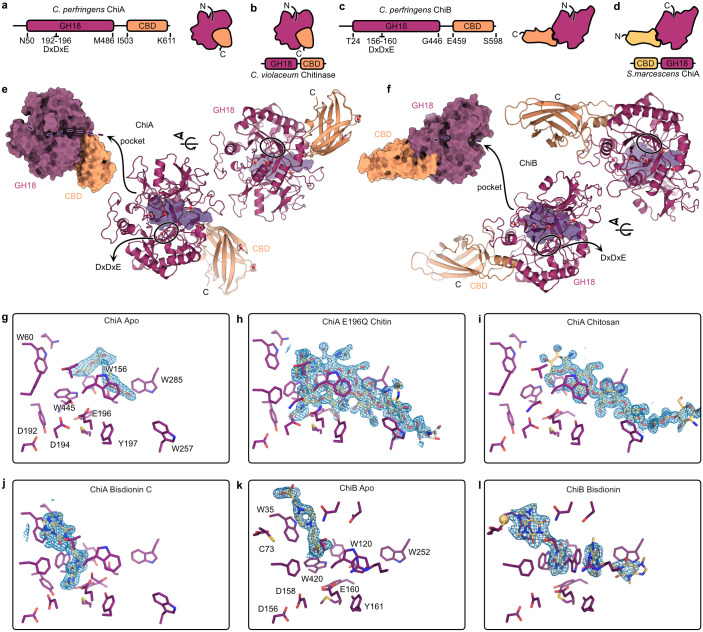
Fig 1. Crystal structures of chitinases ChiA and ChiB.
(A, C) Schematic overview of protein domain organization and overall structures of ChiA and ChiB, both consisting of an N-terminal glycosyl hydrolase 18 (GH18) Trios-phosphate Isomerase (TIM) barrel followed by a putative carbohydrate binding domain (CBD). (B, D) Comparison of overall protein structures of ChiA and ChiB with those of previously studied chitinases (Chromobacterium violaceum and Serratia marcescens). (E,F) Surface representation of the protein crystal structures of ChiA and ChiB identifying the glycosyl hydrolase 18 domain (purple), carbohydrate binding domain (orange) and their open and tunnel shaped binding cleft, respectively, containing the DxDxE binding motif. The transparent purple volume in the cartoon representation of the crystal structures describes the ligand-binding pocket. (G,H, I, J, K, L) Ligand interactions of ChiA or ChiB with substrates: chitin, chitosan or bisdionin C. Ligand OMIT maps are shown as blue mesh representing the difference electron density Fourier coefficient mFo-DFc contoured at + 3 σ carved 2A around the ligands. Maps were generated by performing reciprocal space coordinate refinement after randomizing all atoms on average 0.4 Å in the absence of the ligands.