Figure 2: A GETomics approach to understanding COPD and other chronic human diseases.
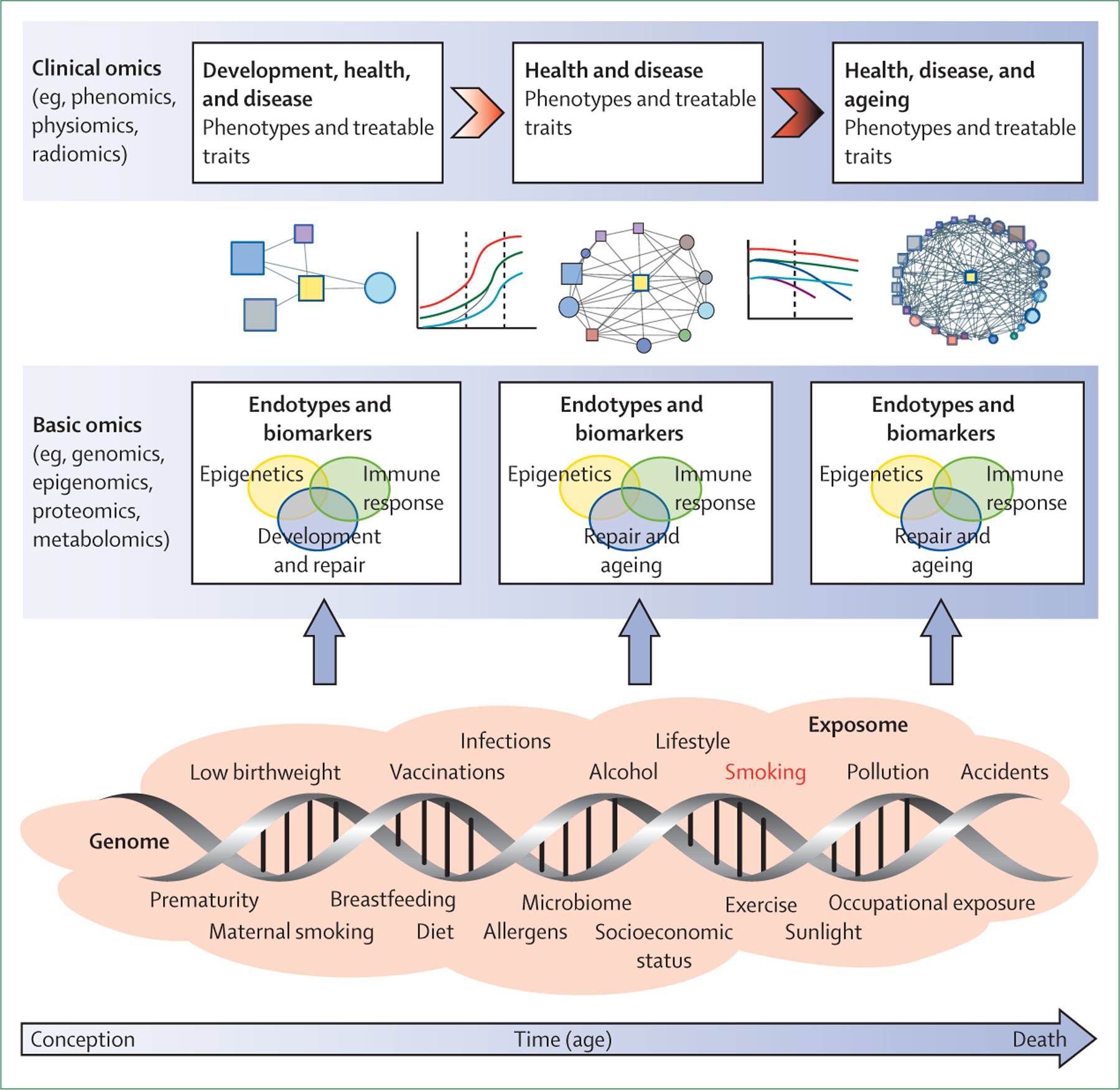
The biological effects and clinical outcomes of different gene–environment interactions depend not only on their specific characteristics, but also on a time dimension—ie, the age of the individual at which the interaction occurs and the cumulative history of the individual’s previously encountered gene–environment interactions. We propose that future research should take a holistic approach that considers the range of interactions between genes (G) and the environment (E) that occur over an individual’s lifespan (time, T) in the context of integrated omics approaches (ie, GETomics) to better understand the pathogenesis of COPD (and probably other chronic human diseases). Examples of environmental factors (the exposome30–33), from conception to death, are represented by orange shading. The positions of different exposures included in the shaded area are not necessarily related to the time axis (arrow) and might occur several times during the lifespan. At different timepoints, these environmental factors interact with the genomic background of the individual through epigenetic and other mechanisms that might be identified through various basic omics approaches. These interactions induce biological responses (endotypes34)—such as innate or acquired immune responses—that modulate organ structure (development, maintenance and repair, ageing) and function. Biomarkers for these endotypes are needed to be able to characterise objectively the pathogenic mechanisms linked to altered lung structure and function. Modulation of organ structure and function, represented here by different lung function trajectories associated with development and ageing, determines long-term phenotypes associated with health and disease, which can be explored through clinical omics approaches. COPD=chronic obstructive pulmonary disease.
