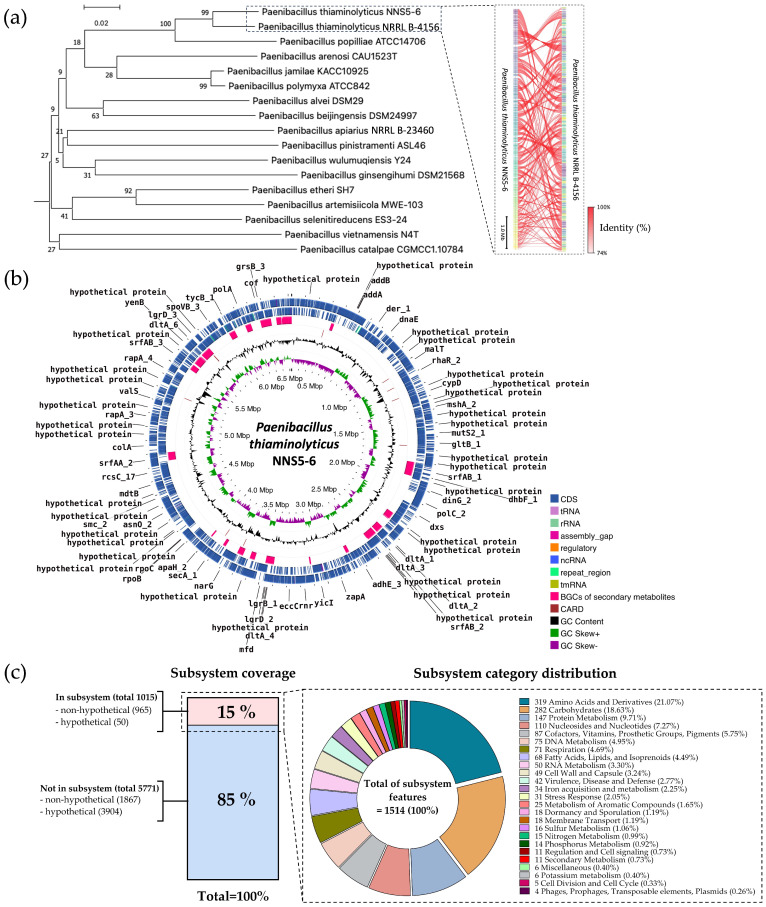
Figure 6.
The genome-based phylogenetic tree of NNS5-6 was constructed using the GBDP method to identify the closest species and strains, via the TYGS web service. The inset visualized by FastANI demonstrates that the DNA fragments in the NNS5-6 genome are similar to the orthologous DNA fragment against the closest related genome, Paenibacillus thiaminolyticus NRRL B-4156 (a). The circular map of the NNS5-6 genome (6.5 Mb) displays the predicted coding sequences relevant to biological processes, including BGCs of secondary metabolites and antibiotic resistance genes (b). The subsystem technology via RAST covers 15% of the predicted subsystems, revealing the cellular machinery. The inset shows the distribution of subsystem categories from the 15% subsystem coverage, identifying 1514 features responsible for the biological processes of the bacterium (c).