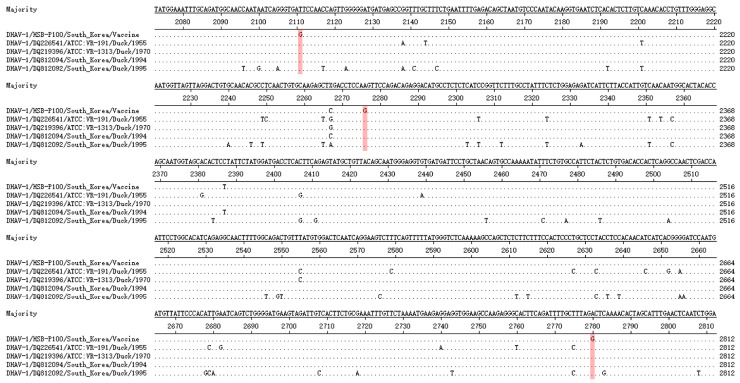
Figure 1.
Multiple sequence alignment between the DHAV-1 vaccine and wild-type strains. The Clustal W alignment method in the MegAlign module of DNASTAR software was used to align the sequences of vaccine strain HSB-P100 with four wild-type strains from ATCC, Korea. Three specific SNPs were found in the vaccine strain at nucleotide positions 2111th, 2276th, and 2780th. The red region indicates these positions in each sequence. “.” indicates nucleotides identical to those in HSB-P100.