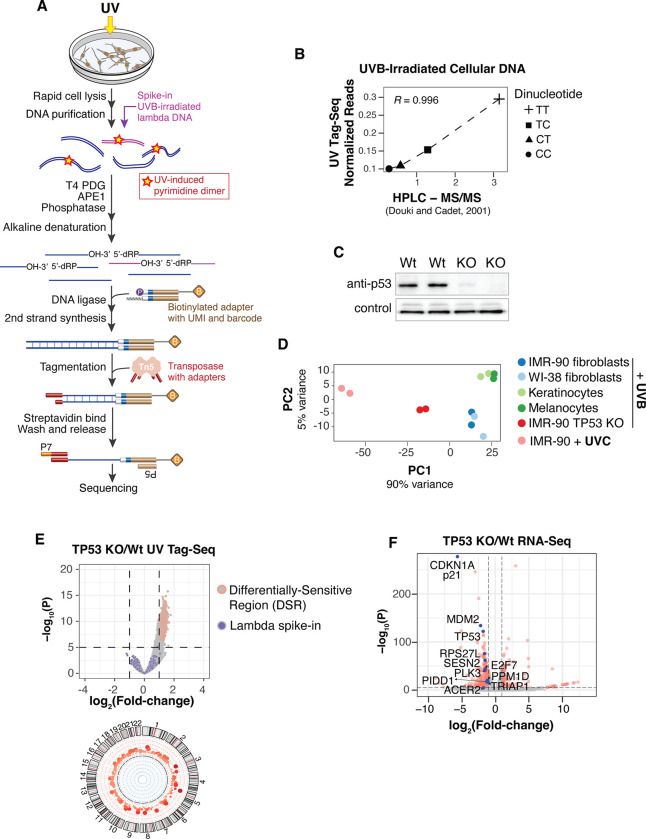
Fig. 1. Loss of p53 increases UV sensitivity in the absence of DNA damage response activation.
(A) Schematic of UV Tag-seq method to quantitatively map UV-induced DNA lesions genome-wide. (B) Correlation of UVB-induced dipyrimidines detected by UV Tag-seq compared with bulk quantification by HPLC-MS/MS (14). (C) Western blot of p53 in wild-type and TP53 knock-out (KO) cells. Technical replicates are shown with loading control (Emerin). (D) Principal Component Analysis (PCA) of UV lesions genome-wide in the indicated cells. (E) (Top) Volcano plot of differential UV sensitivity for TP53 KO cells compared to IMR-90 wild-type cells treated with 200J/m2 UVB. Irradiated lambda DNA used to normalize samples is shown. (Bottom) Circos plot of Differentially Sensitive Regions (DSRs) in autosomes. (F) Volcano plot of RNA-seq comparing TP53 KO and IMR-90 wild-type cells. Red dots denote significantly differentially expressed (SDE) genes. Genes in the “DNA damage response, signal transduction by p53 class mediator” gene ontology pathway are highlighted in blue with corresponding names. Non-SDE genes are not highlighted.