Abstract
A characteristic feature of Alzheimer’s disease (AD) is the formation of neuronal extracellular senile plaques composed of aggregates of fibrillar amyloid β (Aβ) peptides, with the Aβ1-42 peptide being the most abundant species. These Aβ peptides have been proposed to contribute to the pathophysiology of the disease; however, there are few tools available to test this hypothesis directly. In particular, there are no data that establish a dose–response relationship between Aβ peptide expression level and disease. We have generated a panel of transgenic Caenorhabditis elegans strains expressing the human Aβ1-42 peptide under the control of promoter regions of two pan-neuronal expressed genes, snb-1 and rgef-1. Phenotypic data show strong age-related defects in motility, subtle changes in chemotaxis, reduced median and maximum lifespan, changes in health span indicators, and impaired learning. The Aβ1-42 expression level of these strains differed as a function of promoter identity and transgene copy number, and the timing and severity of phenotypes mediated by Aβ1-42 were strongly positively correlated with expression level. The pan-neuronal expression of varying levels of human Aβ1-42 in a nematode model provides a new tool to investigate the in vivo toxicity of neuronal Aβ expression and the molecular and cellular mechanisms underlying AD progression in the absence of endogenous Aβ peptides. More importantly, it allows direct quantitative testing of the dose–response relationship between neuronal Aβ peptide expression and disease for the first time. These strains may also be used to develop screens for novel therapeutics to treat Alzheimer’s disease.
Keywords: amyloid β, C. elegans, neurodegeneration, Alzheimer’s disease, behavioural assays, transgene copy number
1. Introduction
The primary pathological hallmark of Alzheimer’s disease (AD) is the presence of extracellular senile plaques composed of aggregates of fibrillar amyloid β (Aβ) peptides, derived from the proteolytic processing of amyloid precursor protein (APP) [1,2]. The transmembrane protein APP is cleaved sequentially by β-secretase at the N-terminal amino acids 1 and 11, followed by cleavage by γ-secretase, to give rise to a family of Aβ peptides varying between 39 and 43 amino acids in length [3]. One of the most abundant Aβ peptides in the human AD brain is the hydrophobic Aβ1-42 peptide [4,5]. The amyloid cascade hypothesis suggests that the progressive and abnormal accumulation and aggregation of the Aβ peptides in the human AD brain is the primary cause of the disease, and this precedes the other disease symptoms, such as formation of intracellular neurofibrillary tangles, cognitive decline, and dementia [6,7,8]. However, the molecular and cellular mechanisms underlying AD pathology remain contentious and unclear [9,10,11].
Despite genetic evidence and the demonstrated involvement of Aβ in inducing synaptic dysfunction, disrupting neural connectivity, and association with neuronal death in a brain region-specific manner, the amount and distribution of extracellular Aβ deposition are only weakly correlated with the clinical expression and severity of the disease [12]. This weak correlation between the plaque burden and disease severity has been used to criticise the amyloid cascade hypothesis. Furthermore, in many cases the level of the soluble Aβ correlates with the disease burden better than the insoluble Aβ version [13,14]. Additionally, it has been suggested that the soluble Aβ protein is associated with faster cognitive decline, supporting the role of soluble Aβ as a neurotoxic agent of aging [14]. The insoluble Aβ appears only to signify the presence of the disease. Although in physiologic conditions and at a low concentration soluble monomeric Aβ may have a role in neural development and in the regulation of cholinergic transmission [15,16], it appears that Aβ in higher concentrations occurring as oligomers and aggregated forms causes neurotoxicity, impairs blood flow within the cerebral structure, and accelerates neuronal dysfunction [17]. Thus, the exact relationship between Aβ concentration and neuronal dysfunction is unclear, and more studies need to be conducted to clarify it [17].
The nematode Caenorhabditis elegans is an excellent in vivo model system to study the toxicity associated with these Aβ peptides. The simple and compact nervous system of C. elegans consists of 302 neurons. It has well-mapped synaptic connections determined by serial electron micrographs [18,19,20], and a number of robust behavioural phenotypes that have been used to study neuronal function such as chemotaxis, locomotion, egg-laying, pharyngeal pumping, and defecation [21,22,23,24]. The worm is not only capable of displaying a wide range of behavioural phenotypes but also shows behavioural plasticity, including learning behaviours [25].
Several C. elegans strains have been generated previously by expressing the Aβ1-42 peptide in different cell types, and several behavioural phenotypes have been correlated with Aβ1-42 expression. For instance, the C. elegans strain expressing Aβ1-42 peptide in the body wall muscle cells shows a severe, fully penetrant, and age-dependent progressive paralysis phenotype [26]. Since AD is a neurodegenerative disorder, a more disease-relevant strain expressing Aβ1-42 in the nervous system using the pan-neuronal promoter unc-119 was reported and showed reduced lifespan and subtle age-associated decline in behavioural functions such as egg laying, pharyngeal pumping, and abnormal head movements [27]. Another study attempted to show the phenotype associated with Aβ1-42 expression from a single inserted copy of the transgene in a single pair of glutamatergic sensory neurons, the BAG neurons. However, there was no significant behavioural phenotype reported in this strain [28]. Although there are a limited number of C. elegans strains expressing the relevant full-length Aβ1-42 peptide in the neurons, expression of Aβ is associated with subtle behavioural deficits in these strains.
In the present study, we sought to evaluate the effects of varying levels of pan-neuronal Aβ1-42 expression on the onset and severity of different behavioural phenotypes in worms by developing additional transgenic C. elegans strains of Aβ toxicity to achieve variation in Aβ1-42 expression. Differences in transgene expression can be mediated by the promoters integrated with the plasmid, as well as the number of copies of the transgene. We hypothesised that the severity of the disease phenotype correlates with the levels of Aβ expression in these transgenic strains, assuming that transcript concentration is correlated positively with protein translation and/or accumulation. Key behaviours in C. elegans include lifespan, growth rate, egg laying and retention, chemotaxis, and motility. If the level of Aβ expression in the nervous system drives differences in disease severity, then it would be expected that as Aβ expression increases, so does the impact on each of the life and health span measurements as worms get older. The pan-neuronal Aβ1-42-expressing transgenic C. elegans strains described in this study showed varying levels of Aβ1-42 expression and also showed variation in the severity of some, but not all, of the behavioural phenotypes that were measured. Overall, all the Aβ-expressing strains showed reduced longevity, impaired egg-laying, and age-related decline in motility in liquid media, accompanied by subtle defects in chemotaxis and some indications of deficits in neurotransmitter signalling. The phenotypic variation may be correlated with the level of Aβ1-42 transcript abundance.
2. Materials and Methods
2.1. Plasmid Construction
The plasmid pPD49.26 was used as the backbone to construct the pan-neuronal human DA-Aβ1-42 expression plasmids using the pan-neuronal promoters of the snb-1 and rgef-1 gene. The plasmid pPD49.26 contains a synthetic intron and a 3’ UTR region from the unc-54 gene, which are essential elements required for appropriate transgene expression in the worm, as well as three polylinker (MCS) regions for cloning exogenous DNA [29]. The expression plasmid containing the human amyloid β (Aβ) 1-42 gene driven by the pan-neuronal promoter of the snb-1 gene was assembled in two steps. First, the signal peptide and the hu-DA-Aβ1-42 were extracted from the plasmid pCL354. The fragment containing the signal peptide and hu-DA-Aβ1-42 was digested using NheI and SacI from pCL354 [26] and cloned into the MCSII of pPD49.26 to generate the promoter-less plasmid pAB42. This newly constructed promoter-less Aβ1-42-expressing plasmid could be used as a template for future plasmid construction, to drive the expression of Aβ1-42 in specific subsets of neurons using different neuronal promoters. Briefly, pCL354 is an expression plasmid that consists of the peptide (hu-DA-Aβ1-42) driven by the muscle-specific promoter unc-54.
Secondly, a 2 kb region upstream of the snb-1 gene (thought to contain the promoter–enhancer sequence) was amplified from C. elegans wild-type genomic DNA with snb-1 promoter-specific forward and reverse primers that add XbaI (5’) and XmaI (3’) sites, respectively [30]. Immolase enzyme (Bioline, Inc., Cincinnati, OH, USA) was used for amplifying the snb-1 promoter fragment in a 10 μL PCR reaction (1 μL DNA template, 0.5 μL Primer mix (10 μM), 0.4 μL dNTPs, 1 μL Buffer (10X), 0.6 μL MgCl2 solution (50 mM), 0.1 μL Immolase enzyme, 6.4 μL HPLC-grade water) using the following PCR conditions: denaturation at 95 °C for 10 min, 35 cycles of denaturation at 95 °C for 15 s, annealing at 61.2 °C for 40 s, and elongation at 72 °C for 2.5 min, followed by final extension at 72 °C for 5 min. The snb-1 fragment PCR product was digested with XbaI and XmaI and cloned into the complementary XbaI (5’) and XmaI (3’) sites of MCSI of plasmid pAB42, resulting in transgene expression plasmid pSNB1AB42. As a control for appropriate pan-neuronal expression, the same snb-1 promoter fragment was also inserted into the complementary sites XbaI (5’) and XmaI (3’) of the promoter-less GFP-containing plasmid, pPD95.77 (Fire vector kit 1995) [29], resulting in pan-neuronal GFP expression plasmid pSNB-1GFP. An additional non-toxic mouse Aβ1-42 control strain was generated by cloning the snb-1 promoter fragment into the MCSI of the pPD49.26 vector backbone. The mouse Aβ1-42 transgene was extracted from the plasmid pGMC110 using the restriction enzymes NheI and SacI and cloned into the MCSII of the pPD49.26 vector containing the snb-1 promoter fragment in the MCSI.
Next, the promoter region 2670 bp upstream of the rgef-1 gene was amplified from the C. elegans wild-type genomic DNA using the rgef-1 promoter-specific forward and reverse primers using the long-range PCR Go Taq enzyme (Promega Corporation, Madison, Wisconsin, USA) [31]. PCR was performed in 10 µL reactions (1 µL DNA template, 0.5 µL Primer mix (10 µM), 3.5 µL HPLC grade water, and 5 µL GoTaq Master mix (2X)) using the following conditions: initial denaturation at 94 °C for 2 min, 35 cycles of denaturation at 94 °C for 30 s, annealing at 69.7 °C for 30 s, and elongation at 70 °C for 3 min, followed by a final extension at 72 °C for 10 min. The amplified rgef-1 promoter fragment was then digested with PstI and XmaI and cloned into the complementary PstI and XmaI sites of the promoter-less Aβ plasmid pAB42, resulting in the expression plasmid pEXAB42 containing the human Aβ1-42 transgene driven by pan-neuronal promoter rgef-1 (F25B3.3). The rgef-1 promoter fragment was also inserted into the complimentary PstI and XmaI sites of the promoter-less GFP-containing plasmid pPD95.77, resulting in transgene expression plasmid pEXGFP. All clones were verified by Sanger sequencing (Macrogen, Korea). All primers used in the study were synthesised by IDT DNA Technologies (Supplementary Table S1).
2.2. C. elegans Maintenance and Strain Details
Standard protocols were employed for the maintenance of C. elegans strains [32]. All worm strains were cultured on standard nematode growth media (NGM) plates seeded with E. coli OP50 culture at 20 °C, unless otherwise stated [33]. The worm strains used in this study were WG731 (transgenic integrated mCherry control strain) (wgIs1[pAV1944(myo-2p::mCherry)]), WG643 (transgenic integrated neuronal Aβ strain) (wgIs2[pAV1944(myo-2p::mCherry)+ pSNB-1AB42 (snb-1p::huAβ1-42)]), WG663 (transgenic integrated neuronal Aβ strain) (wgIs3[pAV1944(myo-2p::mCherry) + pEXAB42(rgef- 1p::huAβ1-42)]), WG625 (transgenic integrated neuronal GFP strain) (wgIs4[pEXGFP(rgef-1p::GFP)]), WG700 (transgenic integrated neuronal GFP strain) (wgIs5[pSNB-1GFP(snb-1p::GFP)]), GRU101 (transgenic integrated YFP control strain) (gnaIs1[myo-2p::YFP]), GRU102 (transgenic integrated neuronal Aβ strain) (gnaIs2[myo-2p::YFP + unc-119p::huAβ1-42]), and CB1112 (cat-2 (e1112)). The standard Bristol N2 strain was used as the wild-type control.
2.3. Microscopy Details for Imaging the Pan-Neuronal GFP Expressing Strains
Glass microscope slides were prepared with agarose pads by pipetting 300 μL of 2% agarose in M9 buffer (22 mM KH2PO4, 42 mM Na2HPO4, 86 mM NaCl, and 1 mM MgSO4, sterilised by autoclaving) onto the slide and flattening slightly using a coverslip, before allowing the agarose to cool and gel. Once the agarose pad was formed, the coverslip was removed. Several worms were picked on each agarose pad and immobilised by heating the slide on a block at 55 °C for 10 s. The agarose pad was then covered with a coverslip. For imaging WG625 (rgef-1p::GFP) worms, images were acquired using either an Upright microscope Nikon Eclipse Ci equipped with a Nikon DS-U3 digital camera (Nikon Corporation, Tokyo, Japan) or a Zeiss LSM 510(Carl Zeiss Microscopy GmbH, Jena, Germany) laser scanning confocal microscope with a Plan Apochromatic 20X/0.8 NA objective. Excitation was achieved with an argon multiline laser at 488 nm (eGFP) and a DPSS laser of 561 nm (mCherry). A long pass LP575 filter was used for detection of mCherry. Z stacks were obtained using ZEN 2 software (Carl Zeiss Microscopy, GmbH, Jena, Germany). For imaging the WG700 (snb-1p::GFP) strain, an inverted Nikon Ti Eclipse microscope was used. The FITC filter was used to visualise GFP fluorescence (~495 nm) in the worms.
2.4. Generation of Transgenic Integrated Strains
To generate the transgenic C. elegans strains described in this study, pan-neuronal hu-DA-Aβ1-42 expression plasmids were introduced into the C. elegans gonads by microinjection. The Aβ-containing plasmids pEXAB42 (rgef-1 promoter) and pSNB-1AB42 (snb-1 promoter) were micro-injected at a concentration of 25 ng/µL along with the marker (myo-2p::mCherry) plasmid pAV1944 (2.5 ng/μL) and sheared genomic DNA (50 ng/μL). Plasmid pAV1944 (myo-2p::mCherry::unc-54 3’UTR) was used as co-injection marker. pAV1944 expresses mCherry specifically in the pharyngeal muscles [34]. This plasmid was a gift from Gary Silverman at the Washington University School of Medicine in St. Louis (Addgene plasmid #37830; http://n2t.net/addgene:37830; accessed on 1 July 2015, RRIDD: Addgene_37830). The genomic DNA was digested with the 4-base cutting restriction enzyme Sau3AI by incubating the genomic DNA with 1X NEBufferTM r1.1 and 10 units of Sau3AI per microgram of DNA at 37 °C for 1 h, followed by heat inactivation of the enzyme at 65 °C for 20 min (New England Biolabs, Inc.). The progeny from the micro-injected worms were selected on the basis of the pharyngeal mCherry expression. The GFP-containing plasmids pEXGFP and pSNB-1GFP were microinjected along with sheared genomic DNA (50 ng/µL GFP plasmid and 50 ng/µL genomic DNA). For GFP transgenic strains, the progeny were selected based on pan-neuronal GFP expression. No marker was co-injected with pSNB-1GFP and pEXGFP, as the transgene itself expresses GFP, which can be easily viewed via epifluorescence microscopy. All constructs after microinjection yielded 5–7 extrachromosomal transgenic F2 lines. Of these, the line that exhibited more than 80% transmission was selected for the integration experiment.
The micro-injected worms containing the transgene were irradiated using X-rays to allow stable integration of the extrachromosomal arrays into the C. elegans genome. Briefly, one hundred age-synchronous L4-stage hermaphrodites were placed on an unseeded 6 cm NGM agar plate and were irradiated with X-rays (3000–4000 rad) for 4 min using an RS 2000 Biological Research Irradiator (Rad Source Technologies, Inc., Buford, GA, USA). The worms were then transferred to seeded plates and allowed to recover overnight in an incubator set at 15 °C. These X-ray-irradiated adults, also known as P0s, were then allowed to lay eggs. About 400 F1 progeny were picked individually on 6 cm seeded NGM agar plates and analysed for increased transgene transmission in the F2 generation. For an F1 that is heterozygous for an integrated array, the percentage of F2s showing the presence of an integrated array upon self-fertilisation is approximately 75%, and one third of the fluorescent F2 progeny will be homozygous for the integrated transgene array. Accordingly, about eight F2 progeny from each F1 parent showing >75% fluorescent progeny were picked individually on 6 cm NGM plates and were analysed for 100% transmission of the transgene [35]. Approximately four F3 progeny from candidate F2 homozygotes were individually picked to confirm genomic integration of the transgene and to avoid selecting sterile worms. The integrated strain was then outcrossed at least three times to the N2 strain to remove any background mutations introduced during the irradiation procedure. After integration, a total of three integrated lines were isolated for each construct.
2.5. Single Worm Lysis and Copy Number PCR Assay
The copy number of the integrated array was determined by quantitative PCR (qPCR) of single worms for each integrated strain.
Single-worm lysis was performed by picking individual worms into a tube containing 20 µL lysis buffer. Samples were incubated for 16 h at 55 °C followed by 1 h at 85 °C. Worm lysates diluted 10-fold with HPLC-grade water were used as a DNA template for the qPCR of all test samples.
Genomic DNA isolated from GMC296 (using the ISOLATE II Genomic DNA kit from Bioline, Inc.) was used as template DNA to generate standard curves for two PCR reactions, one for the target Aβ1-42 gene and another for the single-copy reference gene, Y45F10D.4, which encodes a putative iron sulphur-containing protein [36]. Standard curves were obtained for the Aβ1-42 gene and Y45F10D.4 by generating a 10-fold dilution series (DNA concentration range: 1 ng to 1 × 10−7 ng) of the template DNA. A no-template negative control was included in all the PCR reactions.
The putative Aβ-containing transgenic strains, selected based on pharyngeal mCherry expression, were analysed for the Aβ1-42 transgene copy number. Average efficiencies obtained from the standard curves were used to normalise the Cq value of the test samples. The relative copy number of the Aβ1-42 gene to reference gene Y45F10D.4 was calculated using the ΔΔCt method [37].
Equation (1): Relative copy number: ΔΔCt method
| (1) |
where Cq = quantitation cycle and E = reaction efficiency.
The qPCR for each sample was performed in duplicate. The relative copy number of Aβ1-42 in each transgenic strain was calculated by averaging the copy number of the Aβ1-42 gene of at least eight worms for that particular transgenic strain. Since all integrated lines had comparable copy numbers, only one line per construct was used for further experiments.
The expression of the Aβ1-42 transgene should be determined by a combination of the transgene copy number (measured by qPCR) and the intrinsic promoter activity (measured as FKPM from RNAseq data available at WormBase (www.wormbase.org; accessed on 23 June 2017). To facilitate comparisons between and interpretation of transcript abundance for the transgenic strains, we used an “index of expected expression” that was simply the arithmetic product of FKPM multiplied by the qPCR copy number.
Equation (2): Index of expected expression
| (2) |
2.6. Total RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated from wild-type and transgenic worms using Trizol (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). The frozen worm pellets were homogenised in 500 µL Trizol and incubated at room temperature for 10 min, to which 0.1 volumes of 1–bromo 3–chloropropane (Sigma) were added, mixed thoroughly, and incubated for another 10 min. The sample was centrifuged at 12,000× g for 20 min at 4 °C. The aqueous phase containing RNA was transferred to a fresh tube and 0.8 volumes of isopropanol (Sigma-Aldrich, Inc., St. Louis, MO, USA) was added. The samples were incubated at room temperature for 15 min and centrifuged at 12,000× g for 15 min at 4 °C. The pellet obtained was washed with 75% ethanol and centrifuged for 5 min at 7500× g. The pellet was then air dried and resuspended in nuclease-free water by heating at 60 °C for 10 min to dissolve the RNA. Following RNA extraction, the samples were treated with DNase I and purified (New England Biolabs, Inc., Ipswich, MA, USA) according to the manufacturer’s instructions to remove any residual genomic DNA contamination. cDNA was synthesised using the Tetro cDNA synthesis kit (Bioline, Inc., Cincinnati, OH, USA) according to the manufacturer’s instructions.
Standard curves for reference genes cdc-42 and Y45F10D.4 were generated using 10-fold serial dilutions of N2 cDNA and for Aβ, GMC296 cDNA was used as a template. RT-qPCR was performed using the same PCR reaction and conditions for qPCR as described above. The real-time quantitative RT-PCR data were normalised by geometric averaging of multiple internal control genes, as previously described, which is a modification of the Pfaffl method [38,39,40,41]. The equation used for calculating the relative gene expression using multiple reference genes is as follows:
Equation (3): Equation for calculating relative gene expression in RT-qPCR.
| (3) |
2.7. Lifespan Analysis
Age-synchronous worm populations were obtained by allowing young adult hermaphrodites to lay eggs for 4–6 h on 6 cm NGM plates seeded with Escherichia coli OP50, then carefully washing the plates to remove all worms while leaving the newly laid eggs behind. The eggs obtained were incubated at 20 °C for three days, and young worms on the first day of adulthood were used for all lifespan assays. On day 3, about 20 worms were picked on each 6 cm seeded NGM plate, for a total of 120 worms used for each assay. The number of live and dead worms were counted. The worms were transferred to fresh plates every day until the end of the reproductive period to avoid overlapping generations and to separate them from the larvae and transferred to new plates every second day during the post-reproductive period. A worm was scored as dead if there was no touch-provoked movement. In addition, the worms that were lost either by crawling off the plate (resulting in drying out) or due to internal hatching (bagging) were scored as censored and not incorporated into the analysis. The assay for each strain was repeated at least three times. Survival curves or Kaplan–Meier (KM) curves were generated and analysed in GraphPad Prism 8 software (GraphPad Software, Inc., La Jolla, CA, USA). Survival curves of the transgenic C. elegans strains described in Section 3.1. The Mantel–Cox (log rank) test was used to determine the difference in the distribution of these survival curves and to determine the p-values. Furthermore, the maximal lifespans for all the strains were analysed by OASIS2 [42] using the modified version of the Mann–Whitney U Test, which determines the differences in the distribution tails of survival data, which affects the lifespan. This test also determines the differences in the proportion of the longevity outliers [42,43,44]. Mortality curves were plotted using the commonly used Gompertz equation [45].
Equation (4): Gompertz equation
| (4) |
The initial mortality rate is A, and G is the exponential mortality rate coefficient. The best-fit values were determined by performing a maximum-likelihood ratio test using the flexsurv package in R [45,46]. Data on lifespan experiments for all biological replicates are summarised in Supplementary Table S2.
2.8. Brood Size Assay
Age-synchronous L4 staged worms (day 2 post hatch) were picked individually to seeded NGM plates and incubated at 20 °C for 24 h. About 5–7 worms per strain were used for each experiment, and the worms were transferred to fresh plates every 24 h until the cessation of egg-laying. Any worms that crawled off the plate or showed internal hatching were removed from the experiment. The plates containing the eggs were incubated at 20 °C for 3 days and then the progeny were counted. The experiment was repeated at least twice for each strain.
2.9. Egg Retention Assay
The egg-in-worm assay was performed according to the protocol described in [47], with a few modifications. Briefly, 15–20 L4-stage worms per strain were picked to seeded NGM plates and incubated at 20 °C for 40 h. One the day of the assay, 10 drops of bleach (2:5 v/v 4.2% NaClO:1N NaOH) were dropped on different locations of a Petri dish lid. Individual worms were transferred into each bleach drop and allowed to disintegrate for about 10 min. This gives enough time for the worm cuticle to dissolve and the eggs to remain. The eggs in utero were counted using a dissecting microscope.
2.10. Chemotaxis Assay
Age-synchronous worm populations were obtained by treating gravid hermaphrodites with alkaline hypochlorite (2:5 v/v 4.2% NaClO:1N NaOH) for 3 min. The reaction was stopped by adding 10 mL of M9 buffer to the sample, followed by washes to remove the residual bleach from the sample. The samples were then placed on a shaker at room temperature for 20 h. The newly hatched L1 larvae in M9 buffer were counted and about 500 L1s were transferred to seeded 9 cm NGM plates. This time point was considered to be day 1. Once the worms reached the L4 stage, the plates were washed, and the worms were transferred to FUdR-containing (25 μM) NGM plates seeded with 10% (w/v) E. coli OP50.
Chemotaxis was performed according to a standard protocol [48] with a few modifications. Adult worms on day 4 and day 8 were used for chemotaxis. The worms were washed off the plates with M9 buffer and transferred to a 15 mL tube. The worms were then allowed to settle, and the supernatant was removed. The washing step was repeated a total of three times to remove all residual bacteria. The worms were not centrifuged, as this could affect chemotaxis. The volatile odorant diacetyl, diluted 1:1000 (1 μL in 999 μL ethanol (100%)), was used as the test odorant for the assay. To set up a chemotaxis assay plate, the volatile odorant (1 μL) was spotted at one end of the plate, about 3 cm from the centre of the plate. The vehicle control (1 μL) (100% ethanol) was spotted on the opposite end of the plate, 3 cm from the centre, similar to the odorant spot. In addition, 5% sodium azide (1 μL) was spotted at the odorant and the control spot to immobilise the worms 15 min prior to the assay. To ensure that the worm motility towards the odorant spot was not an artefact, two plates were set up for each assay by spotting the odorant on the right side of one plate and on the left side of the other plate. The worms in buffer solution were transferred to a small piece of Whatman filter paper (grade 1), which was then inverted onto the assay plate to spot the worms at the centre of the plate. After the worms were transferred onto the agar, the Whatman paper was removed using forceps, taking care to not damage the agar surface. The plates were incubated at 20 °C for 1 h, after which they were scored, and the chemotaxis index was calculated as follows:
Equation (5): Equation for calculating chemotaxis index (CI)
| (5) |
The chemotaxis index (CI) was calculated using Excel 2010 (Microsoft, Inc., Redmond, WA, USA), and the bar graph for the chemotaxis index was plotted in GraphPad Prism 8 software (GraphPad Software, Inc., La Jolla, CA, USA) to allow for comparison between the different C. elegans strains.
2.11. Odorant Preference Associative Learning Assay
The odorant preference learning assay was performed according to the protocol described in [49], with a few modifications. Briefly, age-synchronous worm populations were obtained by bleaching gravid adults. Day 4 adult worms were washed three times with M9 buffer and transferred to an unseeded NGM plate. The worms were conditioned for 2 h by placing 2 µL of odorant (diacetyl) on the lid of the plates. In addition, another group of worms were simultaneously starved in the absence of diacetyl. Moreover, a naïve group of worms was incubated on an NGM plate in the presence of food without any odorant. At the end of the starvation period, chemotaxis assays towards diacetyl were performed on the both the naïve and the conditioned groups as described above in Section 2.9.
2.12. Motility Assays
Worm locomotion was measured on solid and in liquid media. To measure motility on solid media, age-synchronous worms were washed thrice with M9 buffer and placed on unseeded NGM plates (6 cm). The excess buffer was wicked away using a Kimwipe. The worms were allowed to acclimatise on the plates for 5 min, after which worm motility was recorded using an iPhone 7 Plus (Apple Inc., Cupertino, CA, USA) through Labcam from iDu Optics (New York, NY, USA), which is a microscope adaptor for the iPhone. The videos of worm motility on solid media were recorded at 25 FPS and scale of 4.39 μm/pixel, whereas the motility in liquid media was recorded at 25 FPS, 8.8 μm/pixel. Motility in liquid media was measured according to the protocol described in [46], with a few modifications. Briefly, about 10–15 worms were washed at least three times with M9 buffer and transferred to 24-well plates containing 1 mL M9 buffer in each well. The worms were allowed to acclimatise in the liquid for 1 min, after which the thrashing was recorded. Adult worms on day 4 (young), day 8 (middle-aged), and day 12 (old worms) were used for all the motility assays. Video recordings were of approximately 45 s−1 min duration. All the videos were then transferred to the computer, converted to .avi format, and analysed using the WormLab software system version 4.0 (MBF Bioscience, Williston, VT USA). The default track settings were used to analyse the videos, with a few exceptions. The swimming or crawling option was selected depending on the type of video being analysed, and the threshold was adjusted manually for every video.
2.13. Basal Slowing Response Locomotion Assays
The basal and enhanced slowing response assays were performed according to an established protocol, with a few modifications [50]. Each 6 cm NGM assay plate consisted of a ring-shaped bacterial lawn of E. coli strain HB101 with an inner diameter of 1 cm and outer diameter of 3.5 cm. The basal slowing response (BSR) assay was performed on well-fed worms, and the enhanced slowing response (ESR) was measured on starved worms. For BSR, well-fed worms were washed off seeded NGM plates, and half of the worms in the buffer solution were transferred to an unseeded NGM plate. The remaining half were placed at the centre of the bacterial ring in the assay plate. All the excess buffer was removed using a Kimwipe, after which the worms were allowed to acclimatise for 5 min. The worm movement on unseeded and seeded plates was recorded. The videos were recorded for 45 s−1 min. To measure the enhanced slowing response assay, worms were washed off seeded NGM plates and transferred to an unseeded NGM plate. The worms were then allowed to starve for 30 min to 2 h, after which some of the worms from the starved plate were picked to the centre of the bacterial ring on an assay plate and allowed to acclimatise for 5 min. The worm movement of starved worms on food and off food was recorded for 45 s to 1 min. All videos were analysed using the WormLab software. The slowing responses were measured on day 4 (young), day 8 (middle-aged), and day 12 (old) adult worms.
2.14. Statistical Analysis
All data are reported as mean ± standard error of the mean (SEM). For the analysis of age-associated data, we employed two-way ANOVA, with both age and genotype as variables, followed by post-hoc pairwise comparison tests. This approach allowed us to account for the differences in expression levels across the different Aβ strains, ensuring that genotype, as a variable, captured the variations in expression levels. For comparisons between transgenic and control strains with varied expression levels, unpaired Student’s t-tests were used to identify significant differences. All analyses were conducted using GraphPad Prism 8 software. A p-value < 0.05 was considered statistically significant.
3. Results
3.1. Pan Neuronal Aβ1-42-Expressing Strains Show Variation in Copy Number and Expression
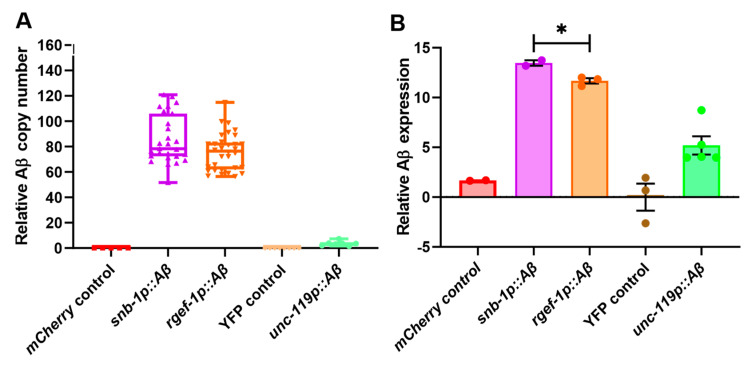
Transgene copy number, which is an important determinant of expression level, was measured by qPCR. After normalisation to a single-copy reference gene, Y45F10D.4, the average relative Aβ1-42 copy number in WG643 (snb-1p::Aβ) was 85.67 ± 3.65 copies per haploid genome, the strain WG663 (rgef-1p::Aβ) contained 75.86 ± 2.58 copies per haploid genome, and GRU102 (unc-119p::Aβ) contained 3.59 ± 0.68 copies per haploid genome (Figure 1A). As expected, the wild-type N2 Bristol, mCherry (WG731), and YFP (GRU101) control strains did not show any copies of the Aβ1-42 transgene.
Figure 1.
Comparison of Aβ copy number and expression levels in the pan-neuronal Aβ strains. (A) Copy number assay showing the number of copies of the integrated Aβ transgene in each strain relative to the reference gene Y45F10D.4 (copy number 1 per haploid genome) (N = 3 replicates, n = 8 worms/replicate). (B) Relative log Aβ expression at the transcript level in the nematodes (mixed life stages), normalised to reference genes Y45F10D.4 and cdc-42 (N = 2–3 replicates, n = 200–300 worms/replicate). An unpaired t-test was used to test for significant differences between Aβ-expressing worms snb-1p::Aβ and rgef-1p::Aβ. * p < 0.05. All bar graph data are reported as mean ± standard error of the mean (SEM).
The expression levels of the Aβ1-42 transgene were measured in these transgenic strains using an RT-qPCR assay and RNA isolated from mixed life-stage cultures (Figure 1B). After normalisation to reference genes Y45F10D.4 and cdc-42, the strain WG643 (snb-1p::Aβ) (high copy number; snb-1 promoter) showed the highest levels of Aβ1-42 expression in mixed life stages (13.47 ± 0.28), followed by WG663 (rgef-1p::Aβ) (11.67 ± 0.26; high copy number, rgef-1 promoter) and GRU102 (unc-119p::Aβ) (5.197 ± 0.91; low copy number, unc-119 promoter). In addition, the relative log-fold expression levels of the human Aβ1-42 transgene in the rgef-1p::Aβ and snb-1p::Aβ expressing strains were measured on day 4 (young), day 8 (middle-aged), and day 12 (old) adults (Supplementary Figure S1A). There was a significant difference in the Aβ1-42 expression level between the rgef-1p::Aβ and snb-1p::Aβ strains in young adults on day 4 (snb-1p::Aβ >> rgef-1p::Aβ), but this difference disappeared in older worms.
We hypothesised that the expression levels of Aβ1-42 in these transgenic strains would depend on a combination of the number of copies of the Aβ transgene integrated into the C. elegans genome and the intrinsic activity transgene promoter. A commonly used measure of promoter activity is fragments per transcript kilobase per million mapped reads from RNAseq data (FKPM). We used comparisons of FKPM values for the native gene in wild-type C. elegans from publicly available RNAseq data as an estimate of the intrinsic strengths of the promoters we used to drive transgenes in the strains described here. The FKPM for the snb-1 gene is 192.8, for rgef-1 it is 4.5, and for unc-119 22.9 (noting that unc-119 expression is not strictly limited to the nervous system) (Supplementary Figure S1B). When considering expression from transgenes, the copy number of the transgene must also be taken into account. This is particularly true for the transgenes described here, all of which we generated by microinjection, which resulted in the formation of extrachromosomal arrays composed of variable numbers of the injected transgenes. Consequently, we calculated an “index of expected relative expression” as the arithmetic product of FKPM × copy number. The observed rank order of expression in either mixed life-stage cultures or in young adults (snb-1p::Aβ > rgef-1p::Aβ > unc-119p::Aβ) was the same as the rank order of expression predicted by the proposed index of expected expression, which combined the intrinsic promoter strength with the transgene copy number (Supplementary Figure S1C).
Additional transgenic C. elegans strains were generated to observe the pattern of transgene expression by expressing GFP driven by the same promoter fragment snb-1 and rgef-1 used for the Aβ constructs. We observed prominent pan-neuronal expression of GFP in both of our imaging experiments, consistent with previous reports characterising snb-1 and rgef-1 expression patterns (Supplementary Figure S1D,E) [30,31]. The GFP signal appeared diffused in the snb-1p::GFP construct because it did not include a nuclear localisation signal. In pan-neuronal GFP expression driven by rgef-1, GFP expression was evident in the head and nerve ring (H & NR), and in both nerve cords and tail neurons (T) (Supplementary Figure S1E).
To summarise, we obtained a panel of pan-neuronal Aβ1-42-expressing strains showing variation in the Aβ expression levels due to a combination of different pan-neuronal promoter activity and variation in the number of integrated Aβ1-42 transgene copies. For easier presentation of the results, we labelled the strains using their promoter and Aβ status: the standard Bristol N2 strain as the wild-type N2 strain, WG643 as snb-1p::Aβ, the WG663 strain as rgef-1p::Aβ, the transgenic control strain WG731 as the pharyngeal-expressed mCherry control, the GRU101 control strain as the YFP control, and the GRU102 strain as unc-119p::Aβ.
Since strain WG643, or snb-1p::Aβ, showed the highest levels of Aβ1-42 expression among the pan-neuronal strains, this strain was expected to show a more severe behavioural defect in comparison to the other strains if Aβ1-42 expression level is the primary determinant of neuronal deficits, particularly in young adults when the expression level differences between the transgenes are strongest. Similarly, the GRU102 strain showed the lowest levels of Aβ1-42 expression, and therefore, it was expected to display more moderate behavioural defects.
3.2. Pan-Neuronal Aβ1-42-Expressing Transgenic C. elegans Strains Show Variation in Lifespan Reduction
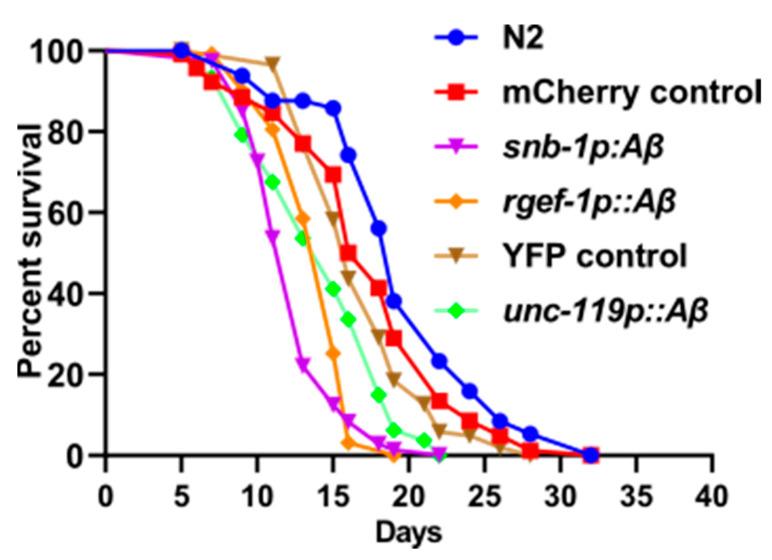
Measurement of lifespan is an initial step to assess whether variation in the expression of the Aβ1-42 transgene in these strains correlates with the severity of the disease phenotype. The transgenic strains snb-1p::Aβ and rgef-1p::Aβ showed a significant reduction in lifespan in comparison to the mCherry control strain (p < 0.0001). The snb-1p::Aβ and rgef-1p::Aβ strains showed a median lifespan of 13.17 ± 0.17 and 14 ± 0.58 (S.E.), respectively, compared to the mCherry control WG731 (17.75 ± 0.25) (Table 1). This is a reduction in median lifespan of 26% for snb-1p::Aβ and 21% for rgef-1p::Aβ. The transgenic strain unc-119p::Aβ also showed a significant reduction in overall lifespan in comparison to the YFP control strain (p < 0.0001). However, the transgenic strain unc-119p::Aβ only showed a slight reduction in median lifespan compared to the relevant myo-2p::YFP (GRU101) control strain (n.s.). The median lifespan of unc-119p::Aβ was 15 days compared to 16 days for the YFP control strain, which is a ~6.25% reduction in median lifespan (Table 1). The differences in median and maximum lifespan of all Aβ expressing strains in comparison to their transgenic controls are listed in Table 1 and survival curves depicted in Figure 2.
Table 1.
Differences in median and maximum lifespan compared to the transgenic control strains.
| Strain Name | Median LS (Mean ± SEM) |
% Change | Maximum LS (Mean ± SEM) |
% Change |
|---|---|---|---|---|
| N2 control strain | 19.3 ± 0.3 | 32.3 ± 0.3 | NA | |
| mCherry control | 17.7 ± 0.3 | NA | 30.7 ± 0.7 | NA |
| snb-1p::Aβ1-42 | 13.2 ± 0.2 | −26% | 19.3 ± 0.3 | −36.9% |
| rgef-1p::Aβ1-42 | 14 ± 0.6 | −21% | 22.7 ± 0.7 | −26.1% |
| YFP control | 16 ± 0.0 | NA | 28 ± 0.0 | NA |
| unc-119p::Aβ1-42 | 15 ± 0.0 | −6.25% | 22 ± 0.0 | −21% |
NA: Not Applicable.
Figure 2.
Transgenic C. elegans strains expressing Aβ peptide in neurons show reduction in life span. Representative Kaplan–Meier survival curves of one biological replicate (n = 120).
The survival data were used to evaluate the differences in the rate of aging between these strains by calculating the initial mortality rate (A) and the Gompertz rate coefficient (G) from the Gompertz equation using maximum likelihood estimation. A lower G value indicates a lower rate of aging. As can be seen in Table 2, there was a significant increase in the Gompertz rate coefficient (G) of the Aβ-expressing strains snb-1p::Aβ and rgef-1p::Aβ in comparison to the mCherry control strain, indicating that the rate of aging in the Aβ-expressing strains was higher than in the control strain. The mortality rate doubling time (MRDT) shows that the chance of worms dying after sexual maturity doubled every 2.16 days for snb-1p::Aβ and every 1.79 days for the rgef-1p::Aβ strain, i.e., rgef-1p::Aβ worms were more likely to die than snb-1p::Aβ worms. In contrast, there was no significant difference in the Gompertz rate coefficient or in the rate of aging between the YFP control strain and unc-119p::Aβ.
Table 2.
Gompertz analysis based on the mortality rate of the Aβ-expressing strains compared to the transgenic controls.
| Strain Name | Initial Mortality Rate (A) | Gompertz Value (G) | Rate of Aging (MRDT) |
|---|---|---|---|
| N2 | 3.65 × 103 | 0.1667 | 4.15 |
| mCherry control | 6.00 × 103 | 0.1677 | 4.13 |
| snb-1p::Aβ1-42 | 3.89 × 103 | 0.3217 ** | 2.16 |
| rgef-1p::Aβ1-42 | 1.91 × 103 | 0.3881 * | 1.79 |
| YFP control | 3.06 × 103 | 0.2274 | 3.05 |
| unc-119p::Aβ1-42 | 3.95 × 103 | 0.2602 | 2.66 |
* p < 0.05, ** p < 0.01.
3.3. Pan-Neuronal Aβ1-42-Expressing Strains Show Defects in Healthspan
Lifespan is a single parameter that measures the amount of time an organism survives. It does not indicate the health (physiological and functional status) of the animal. Health span can be defined as the time that an individual is active, productive, and free from age-associated diseases [51]. Measuring health span, in addition to lifespan, is feasible in C. elegans. C. elegans exhibits several behaviours that are important for its survival and reproduction and are therefore potentially useful indicators of health span because those behaviours are indicative of functionality: locomotion (crawling and swimming), feeding (pharyngeal pumping), defecation, egg-laying, mating, and its ability to sense and respond to chemical, mechanical, and thermal stimuli. Therefore, health span is a concept that attempts to capture the overall state of physical health by measuring complex phenotypes such as reproductive output and coordinated movement. Two such phenotypes in C. elegans are egg-laying rate (eggs laid per unit time) and maximum crawling or swimming speed (on solid or in liquid media, respectively). We sought to test whether variation in the expression of Aβ1-42 influenced the severity of health span indicators such as fecundity and maximum speed.
Reproduction is an excellent indicator of an organism’s general physiological health.
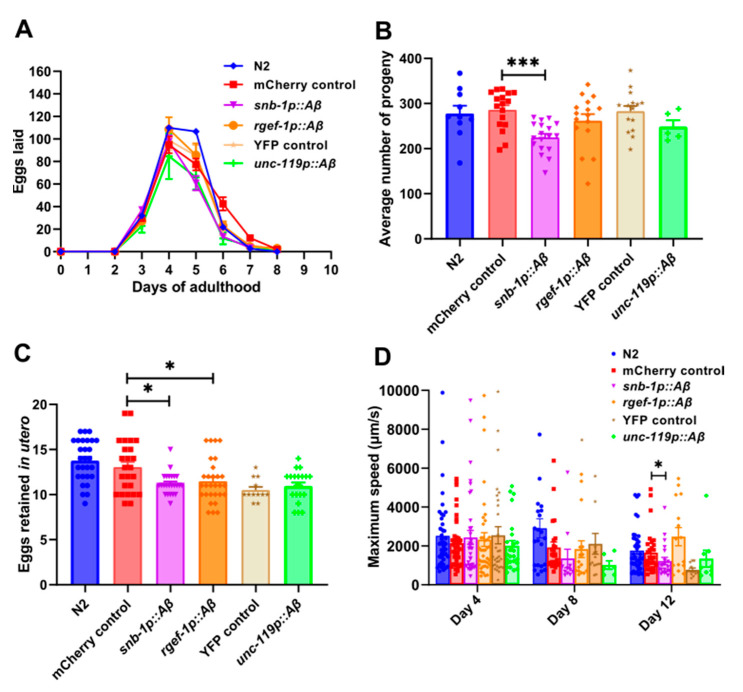
Physiological decline begins earlier when reproduction is adversely affected while the animal is relatively young. In contrast, lifespan assays measure the duration of survival, which reflects the overall health and physiological state of the organism after the reproduction period has ended. Reproductive aging in C. elegans hermaphrodites is characterised by a progressive age-related decline in physiological function beginning on the 5th day and ceasing on the 10th–14th day of adulthood [52,53,54]. Egg-laying is a well-studied aspect in C. elegans physiology and an important reproductive indicator of health span [55]. We sought to measure the egg-laying rate in these transgenic strains to determine the effect of variation in Aβ1-42 expression on reproduction. First, the total number of progeny produced per day during the reproductive span of the worm was counted (Figure 3A) and the total brood size was estimated from these data (Figure 3B). The snb-1p::Aβ strain showed a significant reduction in the number of progeny produced on day 6 (p = 0.0001) and day 7 (p = 0.0034), thereby leading to a significant reduction in the total brood size (225 ± 8.053) in comparison to the mCherry control strain (286 ± 10.37). Although the rgef-1p::Aβ strain showed a significant reduction in the number of progeny produced on day 6 (p = 0.0119) and day 7 (p = 0.0251), there was only a moderate reduction in the total brood size of rgef-1p::Aβ (265 ± 12.44) in comparison to the mCherry control strain WG731. This difference in the total brood size between the rgef-1p::Aβ and mCherry control strain was not statistically significant (p = 0.76). Similarly, the strain unc-119p::Aβ showed a reduction in brood size (248.8 ± 14.72) compared to the YFP control strain (282.8 ± 11.79); however, the difference in the total brood size between the unc-119p::Aβ and the YFP control strain was not statistically significant (p = 0.87).
Figure 3.
Reduction in reproductive output and maximum speed as indicators of health span in Aβ-expressing strains. (A) Number of eggs laid per worm per day during the reproductive span of the animal (n = 3, 5–7 worms/replicate). (B) Total brood size (n = 3, 5–7 worms/replicate). (C) Mean number of eggs retained per worm in utero (n = 3, 15–20 worms/replicate). (D) Maximum speed on solid media (n = 3, 5–15 worms/replicate). An unpaired t-test was used to test for significant differences between Aβ-expressing worms and transgenic controls. ns: not significant, * p < 0.05, *** p < 0.001. All bar graph data are reported as mean ± standard error of the mean (SEM).
Secondly, the number of eggs retained in utero was measured (Figure 3C). Egg laying has a neurological basis, and thus, discriminating between differences in egg production and egg laying is important for assessing whether observed strain variation in brood size is because of changes in the egg-laying neurological circuit [56]. Egg retention reflects a balance between how many eggs are produced and how many are actually laid [47]. The number of eggs retained was significantly reduced in snb-1p::Aβ (p = 0.0192) and rgef-1p::Aβ (p = 0.0419) in comparison to the mCherry control strain, whereas no difference was seen in the Aβ1-42-expressing strain unc-119p::Aβ (p = 0.447) compared to the YFP control strain. Therefore, the ability to lay eggs was not affected but the rate of egg production was reduced, which resulted in an overall reduction in brood size in all the Aβ-expressing transgenic strains in comparison to the transgenic controls.
Additionally, maximum speed on solid media is considered to be an indicator of health span and was measured in these strains [57]. The snb-1p::Aβ did show a significant reduction in maximum speed on day 12 (p = 0.039) by the non-parametric Mann–Whitney test. In contrast, the rgef-1p::Aβ strain did not move faster and the unc-119p::Aβ strain did not show changes in maximum speed relative to the relevant control strain (Figure 3D). Therefore, the higher Aβ-expressing strain snb-1p::Aβ showed a more severe phenotype in comparison to other Aβ-expressing strains for both health span phenotypes that were assessed.
3.4. Pan-Neuronal Aβ1-42-Expressing Strains Show a Significant Reduction in Motility Parameters on Solid and Liquid Media
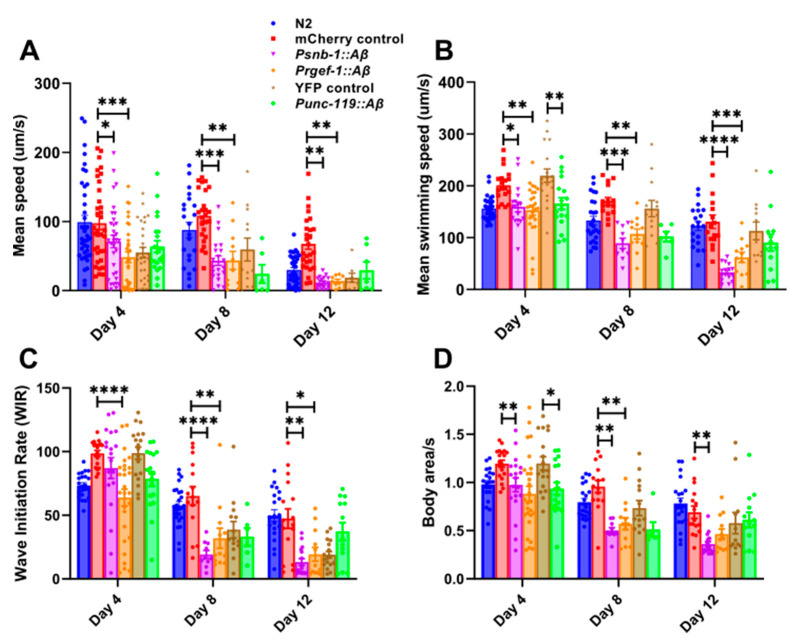
Previous studies have shown that movement on solid and in liquid media measures different aspects of health and behaviour since the kinematics of swimming is distinct from that of crawling [58]. Movement on solid media (crawling) is characterised by a sinusoidal posture, whereas in liquid media, worms show a C-shaped posture [59,60,61]. Several motility parameters in addition to maximum speed were measured in the Aβ-expressing transgenic strains on solid and in liquid media to determine whether differences in the levels of Aβ expression result in variation in specific components of worm motility. Firstly, mean speed on solid media was assessed. There was an age-related decline in the mean speed in all the transgenic strains on solid media (Figure 4A). The snb-1p::Aβ and rgef-1p::Aβ showed a significant reduction in mean speed even in young adult animals on day 4, in addition to day 8 and day 12. Therefore, the moderate and high Aβ-expressing strains showed age-related motility defects on solid media in comparison to the transgenic mCherry control. On the other hand, the low Aβ-expressing strain (unc-119p::Aβ) did not show significant changes in mean speed on solid media in comparison to the YFP transgenic control.
Figure 4.
Pan-neuronal Aβ-expressing strains show age-dependent reduction in motility on solid and in liquid media. (A) Mean speed on solid media (μm/s) (n = 3, 5–15 worms/replicate). (B) Mean swimming speed (μm/s). (C) Wave initiation rate (WIR). (D) Activity in liquid (body area/s) (n = 2, 5–15 worms/replicate). All data analysed by two-way ANOVA followed by post hoc Tukey multiple comparisons test. The snb-1p::Aβ and rgef-1p::Aβ were compared to the transgenic mCherry control strain, and the unc-119p::Aβ was compared to the transgenic YFP control strain. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. All bar graph data are reported as mean ± standard error of the mean (SEM). The genotypes shown in the key in panel A are the same across multiple panels.
Furthermore, motility in liquid was assessed in these strains. The mean swimming speed was significantly lower on day 4 (p = 0.0348), day 8 (p = 0.0001), and day 12 (p < 0.0001) in the transgenic Aβ1-42 expressing strain snb-1p::Aβ compared to the mCherry control strain (Figure 4B). Similarly, the rgef-1p::Aβ strain showed a significant age-related decline in mean swimming speed on day 4 (p = 0.0032), day 8 (p = 0.0025), and day 12 (p = 0.0006) relative to the mCherry control strain. On the other hand, the unc-119p::Aβ strain showed a significant decline in the mean swimming speed only on day 4 (p = 0.0028) in young adults compared to the YFP control strain. The wave initiation rate, which is defined as the number of body waves initiated from the head or tail per minute, was also measured (Figure 4C). There was a rapid decline in the wave initiation rate of the rgef-1p::Aβ on day 4 (p < 0.0001), day 8 (p = 0.0051), and day 12 (p = 0.026) in comparison to the mCherry control strain. However, the snb-1p::Aβ strain showed a rapid decline in the wave initiation rate on day 8 (p < 0.0001) and day 12 (p = 0.0010). In contrast, there was no difference in the wave initiation rate between the YFP control strain and unc-119p::Aβ. The activity index, which is a measure of how vigorously the worm bends, was also analysed. There was an age-related decline in the activity of all the Aβ1-42 expressing strains (Figure 4D). The rgef-1p::Aβ showed a significant decline in activity starting on day 4 (p = 0.0018) in comparison to the snb-1p::Aβ, which showed a reduction in activity on day 8 (p = 0.0005) relative to the mCherry control. In addition, there was a decline in the activity of the unc-119p::Aβ worms on day 4 (p = 0.037), but not on day 8 (p = 0.35) or day 12 (p > 0.9999). Curvature-based motility analysis using parameters such as brush stroke, dynamic amplitude, and curling of the worms was also performed (Supplementary Figure S2). The dynamic amplitude gives a sense of whether the body bends are deep or flat and how much stretching effort occurs in a single body bend (Supplementary Figure S2A). Additionally, the curling parameter determines the percentage of time the animal spends in the bent state and can detect changes in the curl pattern of swimming worms (Supplementary Figure S2B). The low Aβ-expressing strain unc-119p::Aβ showed a significant reduction in curling on day 12 (p < 0.0001) compared to the YFP control strain (Supplementary Figure S2B), which indicates that these worms were experiencing issues with movement control, possibly due to neuromuscular impairment as they aged. Additionally, the brush strokes refer to the sweeping movements made by the worms as they navigate their environment and gives an indication of the depth of the worm movement and extent to which the worms have stretched in a given body bend (Supplementary Figure S2C). There, snb-1p::Aβ in comparison to the transgenic control strain showed a significant reduction in brush strokes on day 12 (Supplementary Figure S2C). A reduction in brush strokes could indicate a decline in exploratory behaviour or underlying issues such as neuromuscular deficits.
The low Aβ-expressing strain unc-119p::Aβ showed a significant decline in swimming parameters, mean swimming speed, and activity compared to the YFP control strain on day 4 (Figure 4B,D) and in the curling parameter in older worms on day 12 (Supplementary Figure S2B). As the decline in motility parameters was more pronounced (relative to the transgenic control) in high and moderate Aβ1-42-expressing strains snb-1p::Aβ and rgef-1p::Aβ but not in unc-119p::Aβ for most of the motility parameters tested, the motility defect was severe in high and moderate Aβ-expressing strains. In summary, all of the Aβ-expressing strains showed an age-related decline in motility parameters in liquid media, but the severity of the motility deficit correlated with the level of Aβ expression.
3.5. Pan-Neuronal Aβ1-42-Expressing Strains Show Defects in Neuronal Function
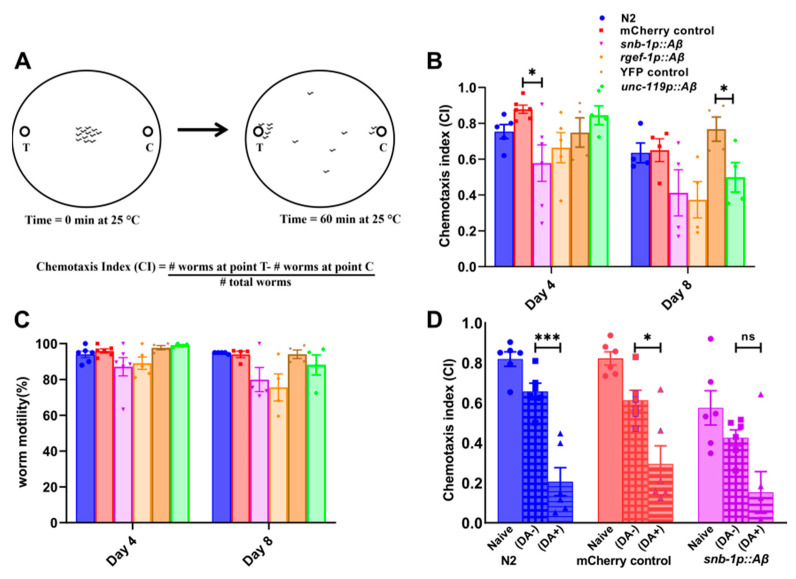
Another behaviour used to assess age-associated neuronal dysfunction is olfaction. Since olfaction in worms is mediated by a specific subset of chemosensory neurons, a well-characterised volatile odorant, diacetyl, was used for chemotaxis assays [48]. This assay was used to measure any differences in the chemotactic responses between these strains as a result of variation in the levels of neuronal Aβ1-42 expression. The chemotaxis index of the worms towards diacetyl was measured in young (day 4) and middle-aged (day 8) adults. In addition, worm mobility was measured on the chemotaxis plates by calculating the percentage of worms that moved away from the origin after 1 h. A schematic illustration of a chemotaxis assay is shown in Figure 5A. Chemotaxis assays measure the fraction of the worms that are attracted to a particular chemical in a given time interval. This fraction is represented as a chemotaxis index, and the values obtained range from +1 to −1, wherein +1 indicates maximum attraction and −1 indicates maximum repulsion.
Figure 5.
Pan-neuronal Aβ-expressing strain shows deficits in chemotaxis and learning. (A) Schematic illustration of a chemotaxis assay. (B) Bar graph showing variation in chemotactic response of Aβ-expressing transgenic C. elegans strains towards volatile odorant diacetyl in young (day 4) and middle-aged (day 8) worms. An unpaired t-test was used to test for significant difference in chemotactic response between transgenic control and Aβ-expressing strains. (C) Percentage motility of young (day 4) and middle-aged (day 8) worms on diacetyl chemotaxis plates. (D) Food-odorant association-based learning assay using volatile odorant diacetyl (n = 6, 100–200 worms per biological replicate). An unpaired t-test was used to test for significant difference in chemotactic response to diacetyl between control and conditioned worms within strains. ns, non-significant, * p < 0.05, *** p < 0.001. All bar graph data are reported as mean ± standard error of the mean (SEM). The genotypes shown in the key in panel B are the same for panel C.
There was a significant decrease in the chemotaxis index of the snb-1p::Aβ strain towards diacetyl on day 4 (CI 0.58 ± 0.10) in comparison to the transgenic control strain (p < 0.01, unpaired t-test) (Figure 5B). Although the snb-1p::Aβ strain also showed a reduction in the chemotaxis index towards diacetyl on day 8 (CI 0.41 ± 0.13) compared to the transgenic control (CI 0.65 ± 0.063), the difference was not statistically significant. In comparison, the transgenic strain rgef-1p::Aβ did show reduced chemotactic ability on day 4 (CI 0.66 ± 0.085) and day 8 (CI 0.37 ± 0.10), although the data were not statistically significant compared to those of the mCherry control strain. Furthermore, the Aβ1-42-expressing strain unc-119p::Aβ also showed a significant reduction in the chemotactic ability towards diacetyl in middle-aged worms on day 8 (CI 0.50 ± 0.082) compared to the transgenic YFP control (CI 0.77 ± 0.07) (p = 0.045, unpaired t-test), but not on day 4 (p = 0.36). There was an age-related decline in the mobility of all the worm strains on day 8, but there was no significant difference between the mobility of the transgenic Aβ1-42-expressing strains in comparison to the controls (Figure 5C). Therefore, the differences in the chemotaxis obtained between the strains were due to chemotactic defects and not due to movement per se.
In summary, the worms moved away from the origin towards the odorant, but in general the chemotaxis indices were not statistically different than those of the relevant controls. The exceptions that were statistically significant were snb-1p::Aβ on day 4 and unc-119p::Aβ on day 8. It is important to note that these mobility data only indicate the ability of the worms to move towards the odorant in a given time period, and do not detect any motility defects between worm strains.
In order to study the effects of Aβ expression on the learning and memory of the worms, food-odorant association-based learning assays were conducted using the high Aβ1-42-expressing strain snb-1p::Aβ. In this assay, young adult worms on day 4 were starved in the presence of the attractant diacetyl for 2 h, and standard chemotaxis assays towards diacetyl were performed after the incubation period. The groups starved in the absence and presence of diacetyl were denoted as diacetyl “−“ and diacetyl “+” in Figure 5D. The naïve group consisted of well-fed worms incubated in the absence of the odorant. The N2 strain and the transgenic mCherry control strain WG731 showed an aversive response by a significant reduction in chemotaxis towards diacetyl after the starvation period. Although the transgenic Aβ-expressing strain group starved in the presence of diacetyl showed a moderate reduction in chemotaxis towards diacetyl after the starvation, the difference between the groups that were starved in the presence and absence of the odorant was not significant.
Moreover, to further validate the severity of the phenotypes observed in our human Aβ-expressing strains, we generated a non-toxic mouse Aβ1-42 transgene-expressing strain driven by same pan-neuronal promoter snb-1. The mouse Aβ1-42 peptide, while capable of aggregation, is significantly less prone to forming the toxic oligomers characteristic of the human Aβ1-42 peptide. The mouse Aβ-expressing strain does not exhibit severe behavioural defects in lifespan, brood size, or motility compared to the human Aβ1-42-expressing strain or the transgenic mCherry control strain (Supplementary Figure S3). This suggests that the toxic phenotypes we observed are indeed specific to the human Aβ1-42 peptide, rather than a general consequence of expressing any aggregating protein or the result of promoter competition.
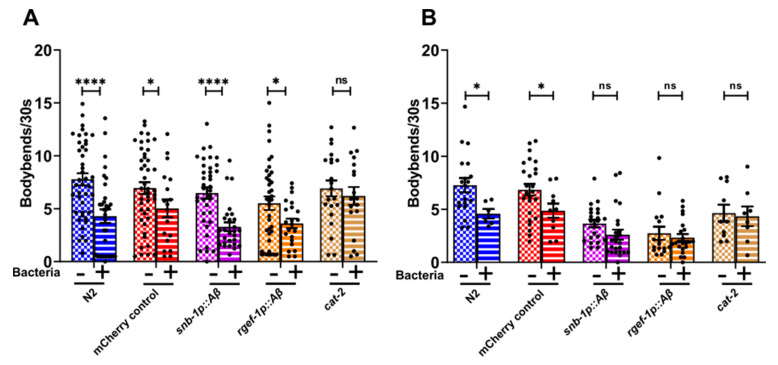
3.6. Pan-Neuronal Aβ1-42-Expressing Strains Exhibit Behavioural Deficits Potentially Linked to Dopaminergic Signalling
Experienced-based locomotion assays are used to study deficits in neurotransmission. The behaviour of worms has been shown previously to change in the presence of food in the environment. For instance, well-fed worms slow down when reintroduced to a bacterial lawn (seeded plate). This slowing response, also known as the basal slowing response (BSR), is experience-based and shown to be dependent on the dopaminergic neuronal pathway [50,62]. The diminished slowing response of well-fed worms on food suggests that there may be impairment in dopaminergic signalling. To determine deficits in dopaminergic signalling, the high and moderate Aβ-expressing strains snb-1p::Aβ and rgef-1p::Aβ were used for the basal slowing response assays. In young adults on day 4, all the transgenic C. elegans strains showed a significant reduction in the number of body bends in the presence of food (seeded plate), except the control strain CB1112 cat-2 (Figure 6A). The strain cat-2 is a negative control strain that carries a loss of function mutation in the cat-2 (tyrosine hydroxylase homolog) gene that inactivates its ability to slow down in the presence of food and is defective in dopamine synthesis.
Figure 6.
Pan-neuronal Aβ-expressing strains show diminished basal slowing response mediated by dopamine. (A) day 4. (B) Day 8. (n = 3, 5–15 worms/replicate). An unpaired t-test was used to test for significant differences in worm movement on seeded and unseeded plates. ns, not significant, * p < 0.05, **** p < 0.0001. All bar graph data are reported as mean ± standard error of the mean (SEM).
On day 8, this slowing response diminished for both the Aβ1-42-expressing strains snb-1p::Aβ and rgef-1p::Aβ in comparison to the mCherry control strain (Figure 6B). These results indicate that there may have been alteration in dopaminergic signalling in the high and moderate Aβ-expressing strains snb-1p::Aβ and rgef-1p::Aβ as the worms aged.
4. Discussion
Although the Aβ peptide has been proposed to play a role in the pathogenesis of AD, direct evidence that quantitatively links Aβ concentration to disease severity is lacking. Low levels of Aβ are protective; however, high levels of Aβ resulting from an imbalance between production and clearance lead to an accumulation of Aβ and an increase in Aβ aggregation, which triggers a pathogenic cascade, ultimately leading to disease [17]. According to this hypothesis, there may be a correlation between Aβ concentration and the initiation, progression, and/or endpoint of the disease. To explore this idea, C. elegans strains expressing varying levels of Aβ1-42 were generated using two different pan-neuronal promoters, rgef-1 and snb-1, and were compared with a previously published strain in which the Aβ1-42 peptide was expressed using the pan-neuronal unc-119 promoter [27]. The unc-119p::Aβ strain was previously shown to have a reduced lifespan, coupled with deficits in egg-laying and locomotory behaviours. The early onset of middle-aged behaviours in these animals correlated with metabolic decline and electron transport failure that preceded Aβ toxicity [27]. The pan-neuronal Aβ1-42 expressing transgenic C. elegans strains described in this study were used to test the hypothesis that the severity of the disease phenotype is correlated with the levels of Aβ1-42 expression. These strains showed varying levels of the Aβ1-42 expression and also showed variation in the severity of some, but not all, of the behavioural phenotypes that were measured. Overall, all the Aβ-expressing strains showed reduced longevity, impaired egg laying, and an age-related decline in motility in liquid media, accompanied by subtle defects in chemotaxis and potential impairment in dopaminergic signalling. In addition, the non-toxic mouse Aβ-expressing strain (snb-1p::mouseAβ) did not show any severe behavioural defects in comparison to the transgenic human Aβ-expressing strain (snb-1p::Aβ) (Supplementary Figure S3). The Aβ peptide found in mice contains three amino acid substitutions within this metal binding domain (Arg5Gly, Tyr10Phe, His13Arg) and significantly decreases the metal binding activity and peroxide production, which suggests that the mouse Aβ is not as intrinsically pathogenic as the human Aβ [63]. These results support the specificity of the observed toxic effects of the human Aβ1-42 peptide.
4.1. Transgenic Aβ1-42-Expressing C. elegans Strains Show Variation in Aβ Expression
The rank order of expression observed amongst the transgenic strains we describe here is the same as that predicted by the index of expected relative expression, i.e., snb-1p::Aβ > rgef-1p::Aβ > unc-119p::Aβ. We propose that the observed differences in transcript abundance give rise to variation in the abundance of Aβ peptide, particularly in younger worms. When comparing the Aβ expression levels by RT-qPCR, there was a 100-fold difference in Aβ expression between snb-1p::Aβ and rgef-1p::Aβ. In addition, rgef-1p::Aβ showed 200% higher Aβ expression than unc-119p::Aβ, which showed the lowest levels of Aβ expression (Figure 1B). That said, our index incorporates the intrinsic promoter strength based on the FPKM value, which is a direct measure of transcript abundance, while Aβ expression as measured by RT-qPCR is a relative measure of transcript abundance and thus dependent on the “housekeeping” transcript(s) that is/are used. In addition, direct comparisons of snb-1p::Aβ and rgef-1p::Aβ strains with the unc-119p::Aβ strain are complicated because the latter was constructed in a different genetic background, has a different transgenic marker that mediates the quantitative variation in Aβ expression, and varies in its intrinsic promoter activity. Another factor potentially influencing the Aβ expression in these transgenic strains is the location of integration sites.
Our categorisation of the transgenic lines as high, moderate, and low expressers is based on expression levels measured in mixed populations of worms. However, these expression levels may diverge with age, potentially influencing the phenotypic outcomes. Additionally, post-transcriptional mechanisms could contribute to the observed differences in expression, independent of promoter activity. Regardless of the quantitative differences in expression (or their cause), if the snb-1p::Aβ strain with the highest levels of Aβ expression shows a more severe behavioural deficit than the rgef-1p::Aβ strain while the unc-119p::Aβ strains shows only subtle behavioural deficits, this supports the hypothesis that levels of Aβ expression correlate with the severity of the disease phenotype, particularly behavioural deficits such as lifespan, motility in liquid, and fecundity.
4.2. Pan-Neuronal Aβ1-42-Expressing Strain Shows Premature Death and Reduction in Fecundity
All the transgenic Aβ1-42-expressing strains showed a significant reduction in median lifespan. The rank order of the reduction in lifespan was the same as the rank order for transcript abundance: snb-1p::Aβ (~ 26%) > rgef-1p::Aβ (~21%) > unc-119p::Aβ strain (~6%) (Table 1). Therefore, the reduction in median lifespan mirrored the levels of Aβ expression. Similarly, the rate of aging, measured by the Gompertz equation, was greater in the higher Aβ-expressing strains snb-1p::Aβ and rgef-1p::Aβ (Table 2) than in the relevant control strain or in the unc-119p:: Aβ strain. However, the unc-119p:: Aβ strain did not age significantly faster than its control. These results indicate that increased levels of Aβ in the C. elegans neurons cause the worms to age faster, whereas low levels of Aβ may influence lifespan without necessarily changing the rate of aging.
In addition, all the Aβ-expressing strains showed a reduction in brood size (Figure 3B). However, this reduction in brood size was only significant for the highest Aβ-expressing strain, snb-1p::Aβ. In addition, there was possibly an impairment in the rate of egg production in these strains, which would result in a reduction in the brood size. That said, reproductive span was not correlated with age-related declines in motility, pharyngeal pumping, or survival probability, which suggests that reproductive span may be regulated independently of these processes [64]. This correlation may change if the hermaphrodite worms are mated with males, which replenishes their sperm, thereby increasing production 3-4-fold and doubling the reproductive span. Thus, although fecundity is accepted as a measure of health span, it may not necessarily correlate with specific age-related neuronal deficits and does not allow for the determination of the underlying causes of change in those measures of health span. It is also important to note that all the young adult Aβ-expressing worms lay eggs at the rate close to the wild type, and the rate of egg production was lower than the transgenic control only on day 6 and day 7 of adulthood. Further experimentation is required to validate these results. For instance, ectopic expression of serotonin to stimulate egg laying could be used to test whether the worm’s response to the treatment restores the egg-laying deficit [65,66]. As the expression of Aβ in the worm nervous system has been shown to impact various physiological functions and its effect on fecundity is not yet fully understood, the observed reduction in fecundity in our Aβ-expressing worms may reflect broader neurodegenerative effects, but further investigation is required to thoroughly establish this relationship.
4.3. Pan-Neuronal Aβ1-42-Expressing Strains snb-1p::Aβ and rgef-1p::Aβ Show a Severe Decline in Motility
Motility has also been suggested as an indicator of health span [57]. There was a rapid age-related decline in several motility parameters, generally in the same rank order of severity as described above for lifespan and fecundity parameters. When comparing motility on solid media, only the high Aβ-expressing strains snb-1p::Aβ and rgef-1p::Aβ showed significant declines in crawling parameters such as mean speed, and only the snb-1p::Aβ strain showed a significant decline in maximum speed, a measure that has been correlated with health span. The low unc-119p Aβ-expressing strain did not show a significant difference in any motility parameter on solid media relative to the control strain. When looking at motility parameters in liquid media, the snb-1p::Aβ and rgef-1p::Aβ strains showed a decline in swimming parameters such as the mean swimming speed, activity, and wave initiation rate (Figure 4). The low Aβ-expressing strain unc-119p::Aβ also showed a decline in all of these swimming parameters, except for the wave initiation rate. Although the wave initiation rate in this strain showed a decline from day 4 to day 8, the change was similar to the one seen in the transgenic control (Figure 4C). It is easier to detect worm motility defects in liquid. Worms crawling on an agar surface encounter mechanical resistance that is 10,000-fold greater than that encountered while swimming in liquid, and this resistance may mask any subtle defects in motility. Despite the higher resistance of crawling on agar versus swimming, swimming in liquid is more energetically demanding than crawling, and there is increase in the metabolic rate of swimming worms [67]. Consistent with the defects in swimming parameters for the unc-119p::Aβ strain, the low Aβ-expressing strain unc-119p::Aβ has been reported to show an ATP deficit at young age [27]. We hypothesise that a more severe energy/ATP generation defect would be observed in the snb-1p::Aβ and rgef-1p::Aβ strains. Since a decline in motility is an early effect of aging, it may eventually compromise other behavioural responses [68].
Locomotion is an important behavioural output in C. elegans regulated by a neural circuit generating sinusoidal bends in response to diverse sensory cues. The sensory cues are integrated by the sensory neurons, which then transmit this information to the interneurons that mediate the flow of this information to the motor neurons and finally to the muscle [20]. Given this context, behaviour is initiated at the neurons upstream of the pathway and executed in the muscle, and therefore, locomotory deficits may arise because of defects in muscle or neurons. For instance, the “uncoordinated” phenotype is an abnormal locomotion phenotype that causes twitching, halting, odd jerky motions, curling or kinking motion, and partial or rigid paralysis, and may arise as a result of mutations in genes expressed in muscles, neurons, or other tissues. There are 111 Unc genes in C. elegans, but 71 of those primarily affect the nervous system and not the muscle [69]. The locomotory deficits observed in the reported strains are neuronal deficits resulting from Aβ expression in the neurons upstream of this pathway. To test this more explicitly, Aβ expression may be targeted to specific neuronal subsets to assess the effects on the severity of the different phenotypes.
4.4. Defects in Chemotaxis and Learning Behaviours Observed in All Aβ1-42-Expressing Strains
Olfaction is affected in AD and is considered to be an early indicator of disease in humans [70]. All the pan-neuronal Aβ1-42 expressing strains showed subtle defects in chemotaxis towards diacetyl. The snb-1p::Aβ and rgef-1p::Aβ strains showed reduced chemotaxis towards diacetyl on day 4, whereas the decline in chemotactic activity was delayed until day 8 in the unc-119p::Aβ strain, demonstrating again that phenotypic severity and/or age of onset are correlated with Aβ expression level. A plausible explanation for this observation is that there is a threshold of Aβ expression at which the phenotype is detectable, and snb-1p::Aβ reaches this threshold for chemotaxis defect earlier because it accumulates Aβ faster. Thus, changes in olfaction are early indicators of age-related cognitive decline due to Aβ in worms, as in humans, because human olfaction is sensitive to the presence of low concentrations of Aβ [70].
High-level cognitive abilities across C. elegans strains are lost much earlier than basic motility and chemotaxis abilities. Therefore, deficiencies in the worms’ learning behaviour can be detected early on [71,72,73,74]. All the memory assays were performed on young adults (day 4) since the chemoreception abilities should be intact on day 4 before the loss of chemotaxis. Food–odour associative learning is a food-dependent behaviour mediated by serotonin, which informs the food status of the environment to the nervous system [75]. The first associative-learning assay used adaptation suppression, as starving wild-type worms in the presence of diacetyl reduces the chemotaxis of the worms towards the odorant. Studies have shown that high and low concentrations of diacetyl induce distinct learning mechanisms [76]. The Aβ-expressing worms showed reduced adaptation suppression when starved in the presence of diacetyl and therefore showed defects in associative learning (Figure 5D). Another study performed the same assay using benzaldehyde and found this associative learning behaviour to be mediated by serotonergic signalling [77]. A similar learning defect was reported for the Aβ3-42-expressing strain when using diacetyl in the learning assay [49].
The nervous system allows the animal to respond flexibly to changes in the environment, which involves alteration of the properties of neurons and synapses caused by the action of neuromodulators such as dopamine and serotonin [50]. Several studies have clearly proven the critical role played by dopamine as a neuromodulator in altering the intrinsic properties within circuits (both pre- and post-synaptically), thereby regulating behaviours such as cognition, egg laying, locomotor activity, and learning [78,79,80,81,82]. The basal slowing response in C. elegans is mediated by dopaminergic neurons that transduce the mechanosensory stimulus (caused by bacteria and surface tension or pressure exerted by the agar surface on the cuticle) through interneurons and motor neurons to the motor circuit, thereby affecting locomotion [50]. In C. elegans, the basal slowing response was sensitive to even small amounts of Aβ: All the Aβ-expressing strains showed a diminished basal slowing response on day 8 (Figure 6B). These findings support the idea that Aβ may contribute to synaptic dysfunction, potentially impacting neurotransmitter deficit signalling. However, the deficits in dopaminergic signalling should be interpreted with caution, as broader neuronal or sensory effects could also be involved (Figure 6). Synaptic dysfunction in AD is considered to be an early hallmark of the disease [49]. These motility-related behaviours are noisy, and experiments to reveal subtle defects that may be the first signs of Aβ toxicity will require large sample sizes. Several studies have reported that serotonergic and dopaminergic neurons are susceptible to aging in humans, specifically in age-related diseases such as AD [83]. To provide a more comprehensive assessment of potential deficits in synaptic transmission, an aldicarb assay could be performed [84]. The aldicarb assay, which evaluates the ability of the neurons to release neurotransmitters in response to a stimulus, could help elucidate whether the observed behavioural deficits are associated with synaptic transmission and could strengthen our understanding of the in vivo toxicity associated with the Aβ species.
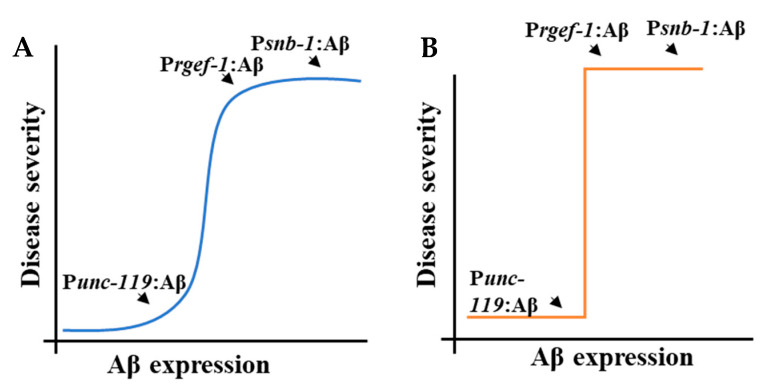
Table 3 shows the ranking of the Aβ-expressing strains according to the severity of the disease phenotype. There were notably statistically significant differences between these strains for some, but not all, phenotypes. Nevertheless, the Aβ-expressing strains were ranked on the basis of phenotype severity, wherein 1 indicates a mild phenotype and 3 indicates a severe phenotype, which is in turn correlated with the level of Aβ transcript abundance. As shown in Figure 7, the relation between Aβ concentration and disease severity could be explained in two ways: (i) a continuous dose–response curve in which is there is gradual increase in disease severity as the concentration of Aβ increases (Figure 7A) or (ii) a threshold response in which there is a range of Aβ concentrations at which there is no phenotype, then a step-wise transition to disease as a threshold concentration is exceeded (Figure 7B). The snb-1p::Aβ and rgef-1p::Aβ strains we describe here had very similar transcript abundance. This means it was not possible to differentiate between a continuously variable concentration-to-phenotype relationship (Figure 7A) and the stepwise threshold (Figure 7B). Strains with intermediate levels of expression are required to investigate this further. It is also possible that different phenotypes will have different relationships between severity and concentration.
Table 3.
Correlation between expression levels and severity of disease phenotype.
| snb-1p:: Aβ1-42 | rgef-1p:: Aβ1-42 | unc-119p:: Aβ1-42 | |
|---|---|---|---|
| Expression level | High | Medium | Low |
| Lifespan | 3 | 2 | 1 |
| Brood size | 3 | 2 | 1 |
| Egg retention | 3 | 2 | 1 |
| Chemotaxis (diacetyl) | 3 | 2 | 1 |
| Motility on solid media | 3 | 3 | 1 |
| Motility in liquid media | 3 | 2 | 1 |
| Basal slowing response | 3 | 3 | NA |
The difference in severity across the various phenotypes is combined with Aβ expression levels in this table to develop an overall phenotype score. Phenotypic severity, combined with expression levels, is color-coded as follows: red for severe, yellow for moderate, and blue for mild. NA: Not Applicable.
Figure 7.
Dose–response curves explaining the correlation between the levels of Aβ expression and the severity of disease. (A) Gradual dose–response curve. (B) Fixed dose–response curve.
It was also observed that some phenotypes, such as deficits in neurotransmitter signalling, were more sensitive to the presence of Aβ. These results therefore provide insight into the relationship between Aβ toxicity and the severity of the behavioural deficit. Future work could include generating several transgenic C. elegans strains expressing levels of the Aβ transgene that span the range between unc-119p:: Aβ1-42 and rgef-1p:: Aβ1-42 to better correlate expression with the severity of the behavioural deficit. This could be achieved by using gene-editing tools such as MosSCI or CRISPR to integrate a specific number of Aβ transgene copies driven by these promoters at a specific location in the C. elegans genome [85]. Alternatively, a titration experiment could be conducted by microinjecting the same promoter construct at different concentrations to generate strains containing varying copies of the Aβ transgene, thereby resulting in a range of Aβ expression. The strains we have reported with differing Aβ expression levels are intended as a starting point for further investigation.
5. Conclusions
This is the first study to demonstrate that varying expression levels using different promoters, and a variable copy number of different integrated arrays establishes a quantitative dose response between Aβ expression and AD-related phenotypes. These data support the conclusion that there is a strong positive correlation between Aβ concentration and the initiation and progression of the disease, rather than the end point of the disease. Therefore, C. elegans serves as a good model to study the relationship between disease progression and Aβ expression because the severity of these behavioural deficits is correlated with the quantitative expression of Aβ. Additionally, these data provide preliminary support for the hypothesis that some behaviours are more sensitive indicators of the disease, such as the basal slowing response assay in which all the Aβ-expressing strains show diminished slowing response irrespective of the Aβ concentration.
Acknowledgments
The authors would like to thank Stephen Doyle for their assistance with the study design and drafting of the manuscript.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells13181598/s1, Table S1: List of primers used in the study.; Figure S1: Variation in pan-neuronal promoter expression chosen for this study; Table S2: Summary of lifespan experiments conducted; Figure S2: Measurement of motility parameters of the transgenic C. elegans strains in liquid media; Figure S3: Molecular and behavioural phenotyping of mouse Aβ-expressing strain in comparison to the human Aβ-expressing strain;.
Author Contributions
Conceptualisation, G.M. and W.N.G.; methodology, N.S. and W.N.G.; validation, N.S., K.G. and W.N.G.; formal analysis, N.S. and W.N.G.; investigation, N.S., K.G. and W.N.G.; resources, W.N.G.; data curation, N.S.; writing—original draft preparation, N.S.; writing—review and editing, N.S., S.M.H., G.M. and W.N.G.; visualisation, N.S. and W.N.G.; supervision, S.M.H. and W.N.G.; project administration, W.N.G.; funding acquisition, W.N.G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research received no external funding. N.S. was supported by an Australian Government Research Training Program (RTP) scholarship.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Glenner G.G., Wong C.W. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984;120:885–890. doi: 10.1016/S0006-291X(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 2.Masters C.L., Simms G., Weinman N.A., Multhaup G., McDonald B.L., Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi R.H., Nam E.E., Edgar M., Gouras G.K. Alzheimer beta-amyloid peptides: Normal and abnormal localization. Histol. Histopathol. 2002;17:239–246. doi: 10.14670/HH-17.239. [DOI] [PubMed] [Google Scholar]
- 4.Haass C., Selkoe D.J. Cellular processing of beta-amyloid precursor protein and the genesis of amyloid beta-peptide. Cell. 1993;75:1039–1042. doi: 10.1016/0092-8674(93)90312-E. [DOI] [PubMed] [Google Scholar]
- 5.Selkoe D.J. SnapShot: Pathobiology of Alzheimer’s disease. Cell. 2013;154:468. doi: 10.1016/j.cell.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Barage S.H., Sonawane K.D. Amyloid cascade hypothesis: Pathogenesis and therapeutic strategies in Alzheimer’s disease. Neuropeptides. 2015;52:1–18. doi: 10.1016/j.npep.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Hardy J.A., Higgins G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 8.Reitz C. Alzheimer’s disease and the amyloid cascade hypothesis: A critical review. Int. J. Alzheimers Dis. 2012;2012:369808. doi: 10.1155/2012/369808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexander A.G., Marfil V., Li C. Use of Caenorhabditis elegans as a model to study Alzheimer’s disease and other neurodegenerative diseases. Front. Genet. 2014;5:279. doi: 10.3389/fgene.2014.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Carlo M. Simple model systems: A challenge for Alzheimer’s disease. Immun. Ageing. 2012;9:3. doi: 10.1186/1742-4933-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schon E.A., Area-Gomez E. Is Alzheimer’s disease a disorder of mitochondria-associated membranes? J. Alzheimers Dis. 2010;20((Suppl. S2)):S281–S292. doi: 10.3233/JAD-2010-100495. [DOI] [PubMed] [Google Scholar]
- 12.Murphy M.P., LeVine H., 3rd Alzheimer’s disease and the amyloid-beta peptide. J. Alzheimers Dis. 2010;19:311–323. doi: 10.3233/JAD-2010-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLean C.A., Cherny R.A., Fraser F.W., Fuller S.J., Smith M.J., Beyreuther K., Bush A.I., Masters C.L. Soluble pool of Abeta amyloid as a determinant of severity of neurodegeneration in Alzheimer’s disease. Ann. Neurol. 1999;46:860–866. doi: 10.1002/1531-8249(199912)46:6<860::AID-ANA8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 14.Yu L., Petyuk V.A., Tasaki S., Boyle P.A., Gaiteri C., Schneider J.A., De Jager P.L., Bennett D.A. Association of Cortical beta-Amyloid Protein in the Absence of Insoluble Deposits With Alzheimer Disease. JAMA Neurol. 2019;76:818–826. doi: 10.1001/jamaneurol.2019.0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harkany T., Abraham I., Timmerman W., Laskay G., Toth B., Sasvari M., Konya C., Sebens J.B., Korf J., Nyakas C., et al. beta-amyloid neurotoxicity is mediated by a glutamate-triggered excitotoxic cascade in rat nucleus basalis. Eur. J. Neurosci. 2000;12:2735–2745. doi: 10.1046/j.1460-9568.2000.00164.x. [DOI] [PubMed] [Google Scholar]
- 16.Sadigh-Eteghad S., Talebi M., Farhoudi M., Golzari S.E.J., Sabermarouf B., Mahmoudi J. Beta-amyloid exhibits antagonistic effects on alpha 7 nicotinic acetylcholine receptors in orchestrated manner. J. Med. Hypotheses Ideas. 2014;8:49–52. doi: 10.1016/j.jmhi.2014.01.001. [DOI] [Google Scholar]
- 17.Sadigh-Eteghad S., Sabermarouf B., Majdi A., Talebi M., Farhoudi M., Mahmoudi J. Amyloid-beta: A crucial factor in Alzheimer’s disease. Med. Princ. Pract. 2015;24:1–10. doi: 10.1159/000369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalfie M., Sulston J.E., White J.G., Southgate E., Thomson J.N., Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J. Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White J.G., Southgate E., Thomson J.N., Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 20.de Bono M., Maricq A.V. Neuronal substrates of complex behaviors in C. elegans. Annu. Rev. Neurosci. 2005;28:451–501. doi: 10.1146/annurev.neuro.27.070203.144259. [DOI] [PubMed] [Google Scholar]
- 21.Croll N.A. Behavioural analysis of nematode movement. Adv. Parasitol. 1975;13:71–122. doi: 10.1016/s0065-308x(08)60319-x. [DOI] [PubMed] [Google Scholar]
- 22.Bargmann C.I., Horvitz H.R. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- 23.Hobert O. Behavioral plasticity in C. elegans: Paradigms, circuits, genes. J. Neurobiol. 2003;54:203–223. doi: 10.1002/neu.10168. [DOI] [PubMed] [Google Scholar]
- 24.Schafer W.F. Genetics of egg-laying in worms. Annu. Rev. Genet. 2006;40:487–509. doi: 10.1146/annurev.genet.40.110405.090527. [DOI] [PubMed] [Google Scholar]
- 25.Byrne J.H., Walker D.S., Chew Y.L., Schafer W.R. Genetics of Behavior in C. elegans. Oxford University Press; Oxford, UK: 2017. [DOI] [Google Scholar]
- 26.McColl G., Roberts B.R., Pukala T.L., Kenche V.B., Roberts C.M., Link C.D., Ryan T.M., Masters C.L., Barnham K.J., Bush A.I., et al. Utility of an improved model of amyloid-beta (Abeta(1)(-)(4)(2)) toxicity in Caenorhabditis elegans for drug screening for Alzheimer’s disease. Mol. Neurodegener. 2012;7:57. doi: 10.1186/1750-1326-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fong S., Teo E., Ng L.F., Chen C.B., Lakshmanan L.N., Tsoi S.Y., Moore P.K., Inoue T., Halliwell B., Gruber J. Energy crisis precedes global metabolic failure in a novel Caenorhabditis elegans Alzheimer Disease model. Sci. Rep. 2016;6:33781. doi: 10.1038/srep33781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinnige T., Ciryam P., Casford S., Dobson C.M., de Bono M., Vendruscolo M. Expression of the amyloid-beta peptide in a single pair of C. elegans sensory neurons modulates the associated behavioural response. PLoS ONE. 2019;14:e0217746. doi: 10.1371/journal.pone.0217746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fire A., Harrison S.W., Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-F. [DOI] [PubMed] [Google Scholar]
- 30.Nonet M.L., Saifee O., Zhao H., Rand J.B., Wei L. Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J. Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L., Fu Y., Ren M., Xiao B., Rubin C.S. A RasGRP, C. elegans RGEF-1b, couples external stimuli to behavior by activating LET-60 (Ras) in sensory neurons. Neuron. 2011;70:51–65. doi: 10.1016/j.neuron.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stiernagle T. Maintenance of C. elegans. WormBook; Pasadena, CA, USA: 2006. pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miedel M.T., Graf N.J., Stephen K.E., Long O.S., Pak S.C., Perlmutter D.H., Silverman G.A., Luke C.J. A pro-cathepsin L mutant is a luminal substrate for endoplasmic-reticulum-associated degradation in C. elegans. PLoS ONE. 2012;7:e40145. doi: 10.1371/journal.pone.0040145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mariol M.C., Walter L., Bellemin S., Gieseler K. A rapid protocol for integrating extrachromosomal arrays with high transmission rate into the C. elegans genome. J. Vis. Exp. 2013;82:e50773. doi: 10.3791/50773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Chen D., Smith M.A., Zhang B., Pan X. Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PLoS ONE. 2012;7:e31849. doi: 10.1371/journal.pone.0031849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao X., Huang X., Zhou Z., Lin X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma Biomath. 2013;3:71–85. [PMC free article] [PubMed] [Google Scholar]
- 38.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoogewijs D., Houthoofd K., Matthijssens F., Vandesompele J., Vanfleteren J.R. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 42.Han S.K., Lee D., Lee H., Kim D., Son H.G., Yang J.S., Lee S.V., Kim S. OASIS 2: Online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget. 2016;7:56147–56152. doi: 10.18632/oncotarget.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C., Li Q., Redden D.T., Weindruch R., Allison D.B. Statistical methods for testing effects on “maximum lifespan”. Mech. Ageing Dev. 2004;125:629–632. doi: 10.1016/j.mad.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Gao G., Wan W., Zhang S., Redden D.T., Allison D.B. Testing for differences in distribution tails to test for differences in ‘maximum’ lifespan. BMC Med. Res. Methodol. 2008;8:49. doi: 10.1186/1471-2288-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yen K., Steinsaltz D., Mobbs C.V. Validated analysis of mortality rates demonstrates distinct genetic mechanisms that influence lifespan. Exp. Gerontol. 2008;43:1044–1051. doi: 10.1016/j.exger.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Bansal A., Zhu L.J., Yen K., Tissenbaum H.A. Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proc. Natl. Acad. Sci. USA. 2015;112:E277–E286. doi: 10.1073/pnas.1412192112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gardner M., Rosell M., Myers E.M. Measuring the effects of bacteria on C. elegans behavior using an egg retention assay. J. Vis. Exp. 2013;80:e51203. doi: 10.3791/51203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bargmann C.I., Hartwieg E., Horvitz H.R. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-H. [DOI] [PubMed] [Google Scholar]
- 49.Dosanjh L.E., Brown M.K., Rao G., Link C.D., Luo Y. Behavioral phenotyping of a transgenic Caenorhabditis elegans expressing neuronal amyloid-beta. J. Alzheimers Dis. 2010;19:681–690. doi: 10.3233/JAD-2010-1267. [DOI] [PubMed] [Google Scholar]
- 50.Sawin E.R., Ranganathan R., Horvitz H.R. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/S0896-6273(00)81199-X. [DOI] [PubMed] [Google Scholar]
- 51.Tissenbaum H.A. Using C. elegans for aging research. Invertebr. Reprod. Dev. 2015;59:59–63. doi: 10.1080/07924259.2014.940470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andux S., Ellis R.E. Apoptosis maintains oocyte quality in aging Caenorhabditis elegans females. PLoS Genet. 2008;4:e1000295. doi: 10.1371/journal.pgen.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes S.E., Evason K., Xiong C., Kornfeld K. Genetic and pharmacological factors that influence reproductive aging in nematodes. PLoS Genet. 2007;3:e25. doi: 10.1371/journal.pgen.0030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendenhall A.R., Wu D., Park S.K., Cypser J.R., Tedesco P.M., Link C.D., Phillips P.C., Johnson T.E. Genetic dissection of late-life fertility in Caenorhabditis elegans. J. Gerontol. A Biol. Sci. Med. Sci. 2011;66:842–854. doi: 10.1093/gerona/glr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maulik M., Mitra S., Bult-Ito A., Taylor B.E., Vayndorf E.M. Behavioral Phenotyping and Pathological Indicators of Parkinson’s Disease in C. elegans Models. Front. Genet. 2017;8:77. doi: 10.3389/fgene.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mignerot L., Gimond C., Bolelli L., Bouleau C., Sandjak A., Boulin T., Braendle C. Natural variation in the Caenorhabditis elegans egg-laying circuit modulates an intergenerational fitness trade-off. Elife. 2024;12:RP88253. doi: 10.7554/eLife.88253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hahm J.H., Kim S., DiLoreto R., Shi C., Lee S.J., Murphy C.T., Nam H.G. C. elegans maximum velocity correlates with healthspan and is maintained in worms with an insulin receptor mutation. Nat. Commun. 2015;6:8919. doi: 10.1038/ncomms9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rollins J.A., Howard A.C., Dobbins S.K., Washburn E.H., Rogers A.N. Assessing Health Span in Caenorhabditis elegans: Lessons From Short-Lived Mutants. J. Gerontol. A Biol. Sci. Med. Sci. 2017;72:473–480. doi: 10.1093/gerona/glw248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mesce K.A., Pierce-Shimomura J.T. Shared Strategies for Behavioral Switching: Understanding How Locomotor Patterns are Turned on and Off. Front. Behav. Neurosci. 2010;4:49. doi: 10.3389/fnbeh.2010.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pierce-Shimomura J.T., Chen B.L., Mun J.J., Ho R., Sarkis R., McIntire S.L. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc. Natl. Acad. Sci. USA. 2008;105:20982–20987. doi: 10.1073/pnas.0810359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vidal-Gadea A., Topper S., Young L., Crisp A., Kressin L., Elbel E., Maples T., Brauner M., Erbguth K., Axelrod A., et al. Caenorhabditis elegans selects distinct crawling and swimming gaits via dopamine and serotonin. Proc. Natl. Acad. Sci. USA. 2011;108:17504–17509. doi: 10.1073/pnas.1108673108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ranganathan R., Sawin E.R., Trent C., Horvitz H.R. Mutations in the Caenorhabditis elegans serotonin reuptake transporter MOD-5 reveal serotonin-dependent and -independent activities of fluoxetine. J. Neurosci. 2001;21:5871–5884. doi: 10.1523/JNEUROSCI.21-16-05871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang X., Atwood C.S., Hartshorn M.A., Multhaup G., Goldstein L.E., Scarpa R.C., Cuajungco M.P., Gray D.N., Lim J., Moir R.D., et al. The A beta peptide of Alzheimer’s disease directly produces hydrogen peroxide through metal ion reduction. Biochemistry. 1999;38:7609–7616. doi: 10.1021/bi990438f. [DOI] [PubMed] [Google Scholar]
- 64.Huang C., Xiong C., Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trent C., Tsuing N., Horvitz H.R. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics. 1983;104:619–647. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waggoner L.E., Zhou G.T., Schafer R.W., Schafer W.R. Control of alternative behavioral states by serotonin in Caenorhabditis elegans. Neuron. 1998;21:203–214. doi: 10.1016/S0896-6273(00)80527-9. [DOI] [PubMed] [Google Scholar]
- 67.Laranjeiro R., Harinath G., Burke D., Braeckman B.P., Driscoll M. Single swim sessions in C. elegans induce key features of mammalian exercise. BMC Biol. 2017;15:30. doi: 10.1186/s12915-017-0368-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Glenn C.F., Chow D.K., David L., Cooke C.A., Gami M.S., Iser W.B., Hanselman K.B., Goldberg I.G., Wolkow C.A. Behavioral deficits during early stages of aging in Caenorhabditis elegans result from locomotory deficits possibly linked to muscle frailty. J. Gerontol. A Biol. Sci. Med. Sci. 2004;59:1251–1260. doi: 10.1093/gerona/59.12.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gieseler K., Qadota H., Benian G.M. Development, structure, and maintenance of C. elegans body wall muscle. WormBook. 2017;2017:1–59. doi: 10.1895/wormbook.1.81.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun G.H., Raji C.A., Maceachern M.P., Burke J.F. Olfactory identification testing as a predictor of the development of Alzheimer’s dementia: A systematic review. Laryngoscope. 2012;122:1455–1462. doi: 10.1002/lary.23365. [DOI] [PubMed] [Google Scholar]
- 71.Herndon L.A., Schmeissner P.J., Dudaronek J.M., Brown P.A., Listner K.M., Sakano Y., Paupard M.C., Hall D.H., Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 72.Kauffman A., Parsons L., Stein G., Wills A., Kaletsky R., Murphy C. C. elegans positive butanone learning, short-term, and long-term associative memory assays. J. Vis. Exp. 2011;49:2490. doi: 10.3791/2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kauffman A.L., Ashraf J.M., Corces-Zimmerman M.R., Landis J.N., Murphy C.T. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 2010;8:e1000372. doi: 10.1371/journal.pbio.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morrison J.H., Hof P.R. Life and death of neurons in the aging brain. Science. 1997;278:412–419. doi: 10.1126/science.278.5337.412. [DOI] [PubMed] [Google Scholar]
- 75.Nuttley W.M., Atkinson-Leadbeater K.P., Van Der Kooy D. Serotonin mediates food-odor associative learning in the nematode Caenorhabditiselegans. Proc. Natl. Acad. Sci. USA. 2002;99:12449–12454. doi: 10.1073/pnas.192101699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bernhard N., van der Kooy D. A behavioral and genetic dissection of two forms of olfactory plasticity in Caenorhabditis elegans: Adaptation and habituation. Learn. Mem. 2000;7:199–212. doi: 10.1101/lm.7.4.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nuttley W.M., Harbinder S., van der Kooy D. Regulation of distinct attractive and aversive mechanisms mediating benzaldehyde chemotaxis in Caenorhabditis elegans. Learn. Mem. 2001;8:170–181. doi: 10.1101/lm.36501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsalik E.L., Hobert O. Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans. J. Neurobiol. 2003;56:178–197. doi: 10.1002/neu.10245. [DOI] [PubMed] [Google Scholar]
- 79.Sanyal S., Wintle R.F., Kindt K.S., Nuttley W.M., Arvan R., Fitzmaurice P., Bigras E., Merz D.C., Hebert T.E., van der Kooy D., et al. Dopamine modulates the plasticity of mechanosensory responses in Caenorhabditis elegans. EMBO J. 2004;23:473–482. doi: 10.1038/sj.emboj.7600057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Omura D.T., Clark D.A., Samuel A.D., Horvitz H.R. Dopamine signaling is essential for precise rates of locomotion by C. elegans. PLoS ONE. 2012;7:e38649. doi: 10.1371/journal.pone.0038649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li J., Li D., Yang Y., Xu T., Li P., He D. Acrylamide induces locomotor defects and degeneration of dopamine neurons in Caenorhabditis elegans. J. Appl. Toxicol. 2016;36:60–67. doi: 10.1002/jat.3144. [DOI] [PubMed] [Google Scholar]
- 82.Ezcurra M., Tanizawa Y., Swoboda P., Schafer W.R. Food sensitizes C. elegans avoidance behaviours through acute dopamine signalling. EMBO J. 2011;30:1110–1122. doi: 10.1038/emboj.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mattson M.P. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mahoney T.R., Luo S., Nonet M.L. Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat. Protoc. 2006;1:1772–1777. doi: 10.1038/nprot.2006.281. [DOI] [PubMed] [Google Scholar]
- 85.Nance J., Frokjaer-Jensen C. The Caenorhabditis elegans Transgenic Toolbox. Genetics. 2019;212:959–990. doi: 10.1534/genetics.119.301506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article and Supplementary Materials.