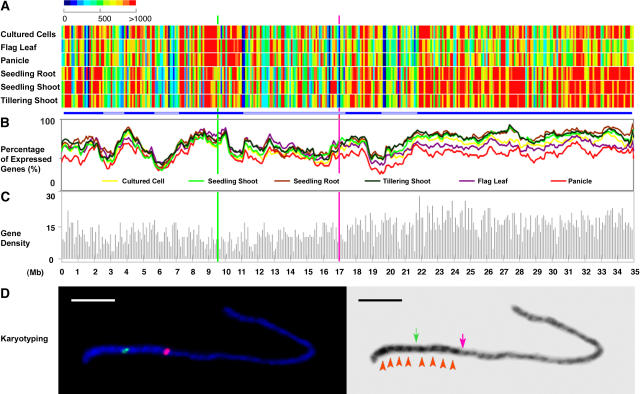
Figure 6.
Chromosomal Transcriptional Analysis of the Nonredundant Protein-Coding Gene Models of Rice Chromosome 4.
(A) Average expression intensities of transcribed fragments in each 100-kb window along the chromosome in each organ or cultured cells. Bars at bottom highlight chromosomal domains based on expression.
(B) Percentage of expressed gene models in a 100-kb window along the chromosome in each organ or cultured cells.
(C) Annotated nonredundant protein-coding gene model density along the chromosome in a 100-kb window.
In (A) to (C), the position of the centromere is represented by a green line, whereas the last major knob of the heterochromatin half of chromosome 4 (as defined in [D] below) is shown by a pink line.
(D) Karyotyping of rice pachytene chromosome 4 and positioning of the euchromatin and heterochromatin border region. Left, chromosome fluorescence in situ hybridization using the centromeric probe CentO (green signal) and a BAC clone, OSJNBa0034E24 (pink signal), located at 16.9 Mb. This BAC clone is located within the last major knob of the heterochromatin half of chromosome 4 and thus is close to the border of the heterochromatin and euchromatin domains. Right, the 4′6-diamidino-2-phenylindole dihydrochloride–stained chromosome 4 at left was converted to a black-and–white image to enhance the visualization of the distribution of euchromatin and heterochromatin. The centromere and euchromatin/heterochromatin border region are indicated by green and pink arrows, respectively. Eight heterochromatin knobs are highlighted by red arrowheads. Bar = 5 μm.