Figure 1.
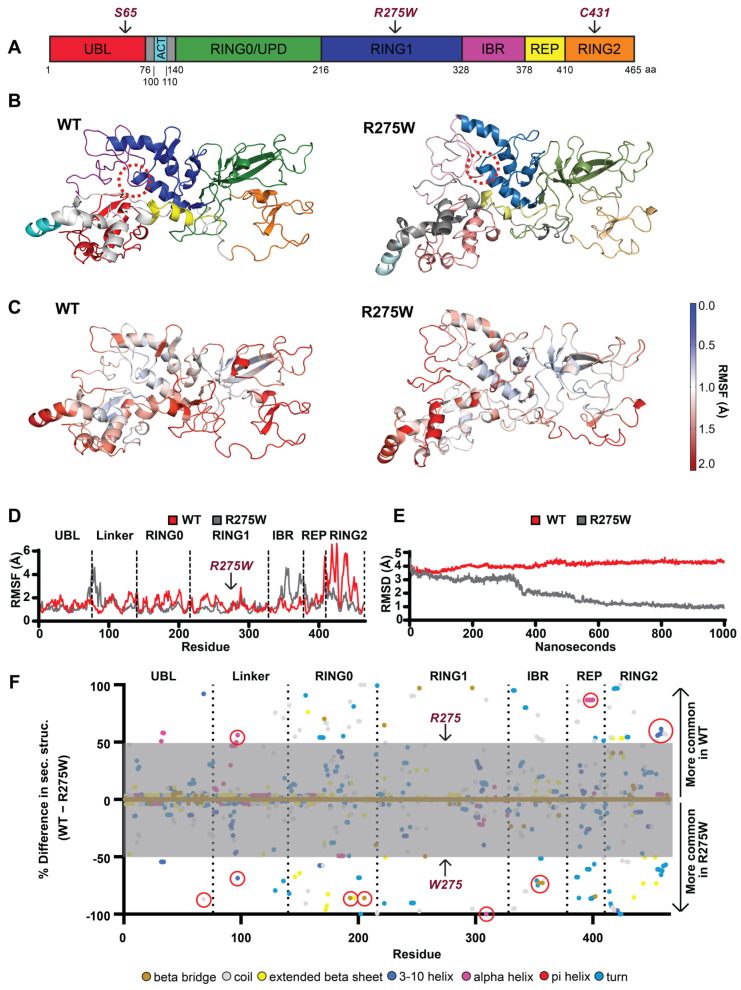
Structure of PRKN WT and p.R275W and conformational changes. (A) Schematic overview of the PRKN protein structure and individual domains: Ub-like domain (UBL, red), linker domain (gray), activating element (ACT, cyan), really interesting new gene domain (RING0, green), RING1 (blue), in-between-RING domain (IBR, purple), repressor element of PRKN (REP, yellow), and RING2 (orange). (B) A 3D model of the autoinhibited structure of PRKN WT and p.R275W with individual domains colored as in (A); the location of position 275 is highlighted by a red dashed circle. (C) A 3D heat map illustration of the root mean square fluctuation (RMSF) for PRKN WT and p.R275W over the course of the simulations. (D) Comparison of RMSF for each residue of PRKN WT (red) and PRKN p.R275W (gray). Domains are labeled and separated with dotted lines. (E) Root mean square deviation (RMSD) of PRKN WT (red) and PRKN p.R275W (gray) structure over 1 µs simulation. (F) Per-residue difference plot of the secondary structure averaged over the entire simulation and colored by secondary structure feature. Shown are percent occupancy times of each residue in a specific secondary structure for PRKN WT (top) and PRKN p.R275W (bottom). Differences greater than 50% (outside the gray area) are considered most meaningful. Secondary structure colors are indicated at the bottom of the figure.