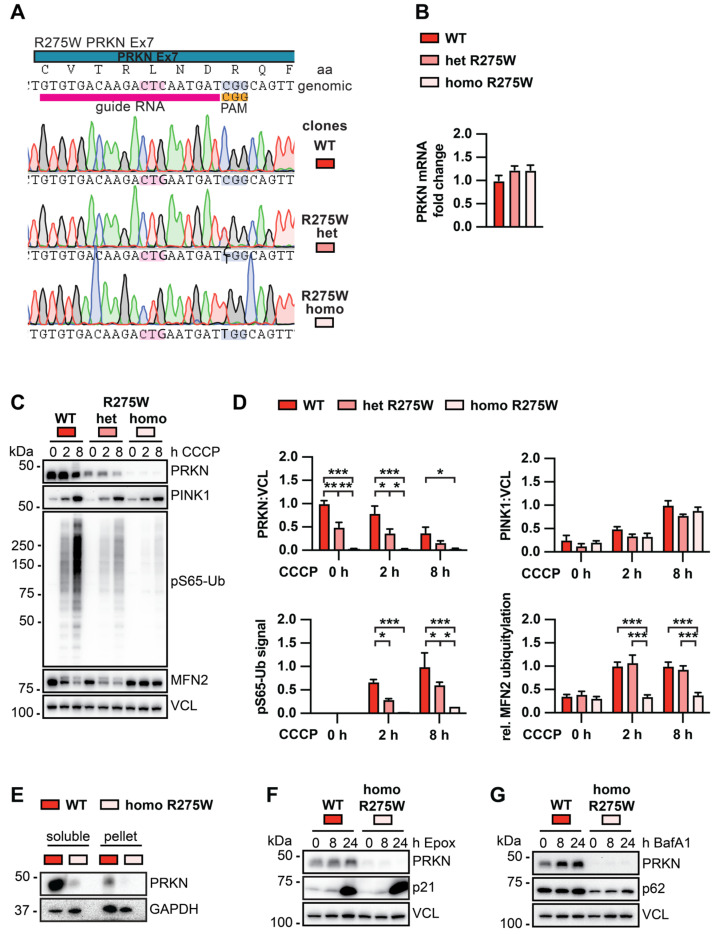
Figure 3.
Biochemical analysis of PINK1-PRKN signaling in isogenic DA neurons with or without the PRKN p.R275W mutation. (A) Schematic of CRISPR–Cas9 target location, gRNA, and results from Sanger sequencing of isogenic clones. (B) PRKN mRNA levels in WT, p.R275W heterozygous, and p.R275W homozygous gene-edited DA neurons. Data shown are mean ± SEM (n = 3). (C) WT, p.R275W heterozygous, and p.R275W homozygous gene-edited DA neurons were treated with 20 µM CCCP for 0, 2, or 8 h. Cell lysates were collected for Western blot analysis and probed with antibodies against PRKN, PINK1, pS65-Ub, MFN2, and VCL. Representative Western blot shows levels of PRKN, PINK1, pS65-Ub, and MFN2 for all three cell lines and treatments. (D) Densitometric analysis of PRKN and PINK1 normalized by VCL. Quantification of pS65-Ub protein levels by sandwich ELISA. Relative modification of MFN2 was calculated as the ratio of the upper (ubiquitylated) to lower (unmodified) MFN2 band. Statistical analysis was performed via two-way ANOVA (* p < 0.05, ** p < 0.01, *** p < 0.001) and data shown are mean ± SEM (n = 3). Only statistically significant comparisons are indicated. (E) Proteins from WT and PRKN p.R275W homozygous gene-edited DA neurons were sequentially extracted in two fractions for solubility analysis and probed with antibodies against PRKN and GAPDH. (F) Gene-edited DA WT neurons and PRKN p.R275W homozygous DA neurons were treated with 200 μM epoxomicin for 0, 8, or 24 h to test PRKN degradation via the proteasome. Cell lysates were collected for Western blot analysis and probed with antibodies against PRKN, p21, and VCL. (G) Gene-edited DA neurons were treated with 400 μM bafilomycin A1 for 0, 8, or 24 h to test for PRKN levels upon inhibition of the autophagosome-lysosome fusion. Cell lysates were collected and probed against PRKN, p62, and VCL.