Table 1.
Chemical structure and physicochemical properties of relevant HsrA ligands identified by the fluorescent thermal shift-based high-throughput screening (HTS) of the Maybridge HitFinderTM chemical library.
| Ligand | Chemical Structure | Formula | Molecular Weight (Da) | Log P 1 | H-Bond Donors | H-Bond Acceptors | Lipinski Violations | TPSA (Å) 2 | Rotatable Bonds | Log D 3 |
|---|---|---|---|---|---|---|---|---|---|---|
| I |
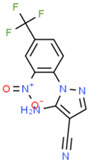
|
C11H6F3N5O2 | 297.19 | 1.56 | 1 | 7 | 0 | 113.45 | 3 | 2.26 |
| IV |
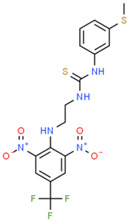
|
C17H16F3N5O4S2 | 475.47 | 3.08 | 3 | 7 | 0 | 185.12 | 11 | 4.97 |
| V |
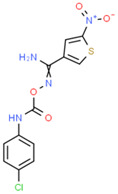
|
C12H9ClN4O4S | 340.74 | 1.99 | 2 | 5 | 0 | 150.77 | 6 | 2.92 |
| VIII |
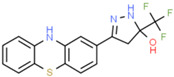
|
C16H12F3N3OS | 351.35 | 3.50 | 3 | 5 | 0 | 81.95 | 2 | 3.60 |
| XI |
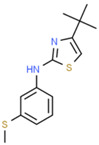
|
C14H18N2S2 | 278.44 | 4.18 | 1 | 1 | 0 | 78.46 | 4 | 4.77 |
| XII |
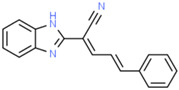
|
C18H13N3 | 271.32 | 3.53 | 1 | 2 | 0 | 52.47 | 3 | 3.79 |
Molecular properties obtained from the SwissADME web tool “http://www.swissadme.ch/ (accessed on 19 September 2024)” and the ChemSpider database “www.chemspider.com (accessed on 19 September 2024)”. 1 Log P values correspond to consensus Log Po/w from SwissADME. 2 TPSA, topological polar surface area. 3 Log D values correspond to ACD/LogD (pH 7.4) from ChemSpider.
