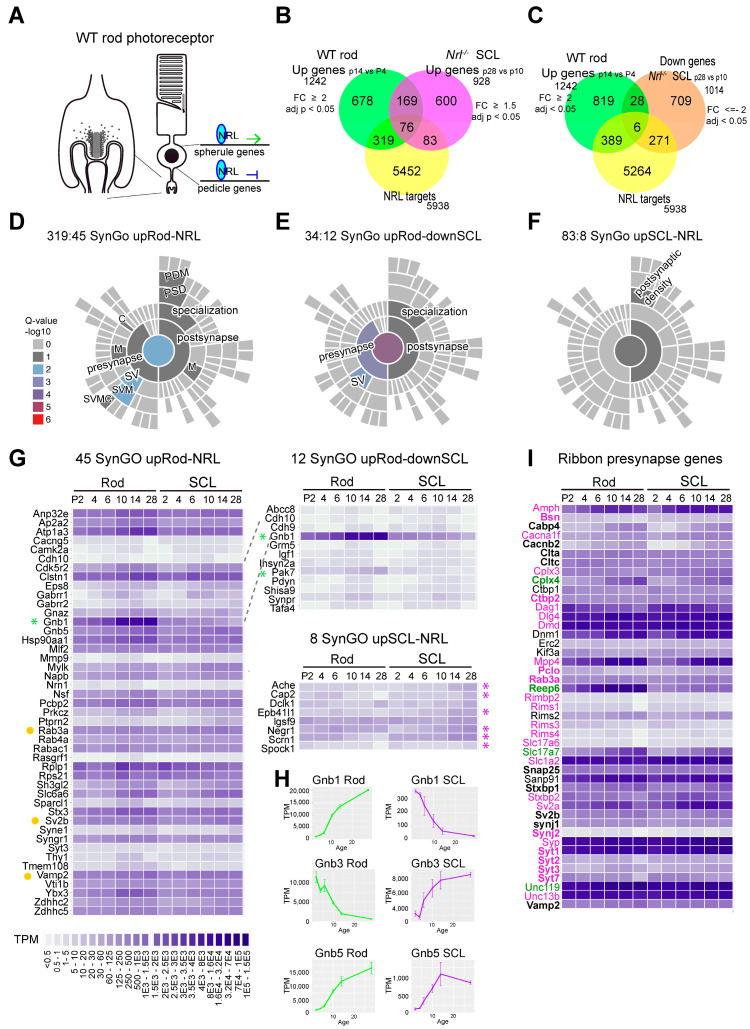
Figure 7.
RNA-seq and CUT&RUN-seq analyses of photoreceptor pre-synapse genes. (A) Schematic drawing of NRL gene regulation in rod photoreceptors. NRL activates rod genes and suppresses cone genes. (B,C) Venn diagrams displaying differently expressed genes in rods, SCLs, and NRL-binding genes. (D–F) SynGO visualizations of upRod-NRL, upRod-downSCL, and upSCL-NRL. (G) SynGO synapse genes, in order from P2 to P28 in transcript per million (TPM) heatmaps: SynGO upRod-NRL (left), SynGO upRod-downSCL (upper right), SynGO upSCL-NRL (down right). Genes in which transcripts are upregulated in developing rods and down-regulated in developing SCLs up to P28 are indicated with a green asterisk, and genes in which transcripts are down-regulated in rods and upregulated in developing SCLs are indicated with a purple asterisk. Known ribbon-associated genes are indicated with a yellow dot. (H) Gnb1, Gnb3, and Gnb5 expression in developing rods and SCLs measured by RNAseq dataset (TPM). The line plots (average ± SD) from all relevant transcripts in 2 to 4 bio-replicates were generated using ggplot2 from R studio. (I) Ribbon synapse genes, in order from P2 to P29 in the TPM heatmap. Gene names in magenta correspond to genes showing higher expression in SCLs than rods at P28. Green gene names correspond to genes showing higher expression in rods than SCLs at P28. Gene names in bold font indicate that NRL binds to the genes. Abbreviations: WT, wild-type; Nrl, neural retina leucine zipper; upRod-NRL, upregulated NRL binding genes in developing WT rods; upSCL-NRL, upregulated NRL binding genes in developing S-cone-like photoreceptors; downSCL, down-regulated genes in developing S-cone-like photoreceptors; adj. p, adjusted p-value; TPM, transcript per million; SynGO, synaptic Gene Ontologies and annotations.