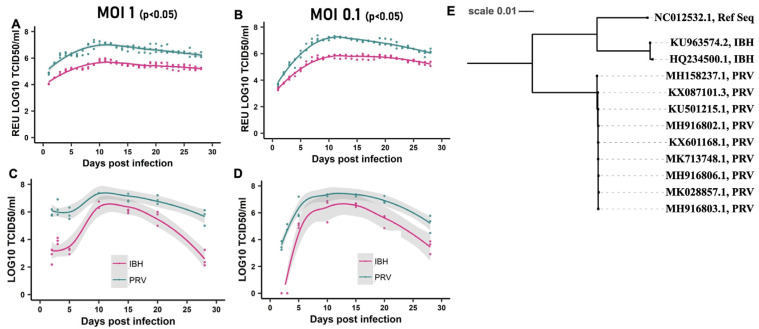
Figure 1.
ZIKV persists in CHME-3 cells: (A,B) Quantification of Zika virus RNA produced de novo in CHME-3 cells by African lineage ZIKV (ZIKV-IBH) or Asian Lineage ZIKV (ZIKV-PRV) over 28 days after infection at a multiplicity of infection 0.1 or 1. Quantification obtained by qRT-PCR represented as relative equivalent units of TCID50. All values are presented as mean ± SEM, n = 3 replicates. (C,D) ZIKV-infectious titer produced de novo in CHME-3 cells by African lineage ZIKV (ZIKV-IBH) or Asian Lineage ZIKV (ZIKV-PRV) over 28 days after infection at a multiplicity of infection 0.1 or 1 at selected time points. Quantification obtained by TCID50. (E) Phylogenetic analysis of complete coding regions of the Asian and African ZIKV strains. Maximum likelihood tree based on Tamura–Nei model with bootstrap support of 100 replicates constructed using the Geneious Prime® 2022.1.1 software, drawn to scale. Viral strains are shown with their NCBI GenBank ID and lineage information. All values are presented as mean ± SEM, n = 3 biological replicates.