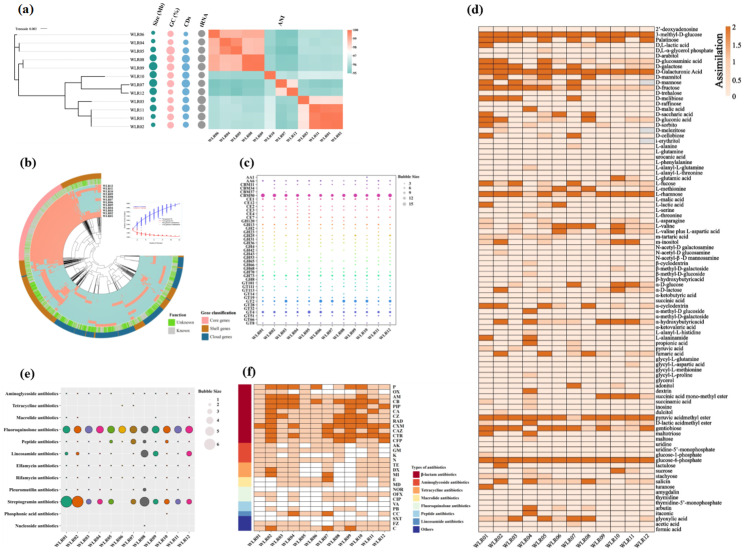
Figure 1.
Comparative genome analysis and association of geno- and phenotypes of Lm. reuteri strains. (a) Phylogenetic tree constructed with the neighbor joining method based on core gene alignment (left). Genome features include genome sizes, GC%, CDs numbers, number of tRNA, and average nucleotide identity (ANI) (right). (b) Pan- and core-genome analysis showing the growing pan-gene numbers as genome numbers increase (top right); pan-genome analysis of Lm. reuteri strains. Each ring represents one strain, and each radial extension in the ring corresponds to a particular gene (red: present; blue: absent). The outer grey–green ring represents COGs functionality annotation, with grey indicating known and green indicating unknown functions. The outermost ring highlights particular gene collections: core, pink; shell genes, brown; and cloud genes, dark blue. (c) Genotypic and phenotypic analysis of carbohydrate utilization, genes related to carbohydrate activity enzymes. (d) BIOLOG test results for carbon sources assimilation. (e) Genome annotation for antibiotic resistance. (f) Experimental observation on bacterial resistance to 30 antibiotics (white = tolerant; faint yellow = intermediate; yellow = intolerant); the Lm. reuteri strain WLR08 was analyzed in parallel for reference. Abbreviations: P: penicillin; OX: oxacillin; AM: ampicillin; CB: carbenicillin; PIP: piperacillin; CA: cephalexin; CZ: cefazolin; RAD: cephradine; CXM: cefuroxime; CAZ: ceftazidime; CTR: ceftriaxone; CFP: cefoperazone; AK: amikacin; GM: gentamicin; K: kanamycin; N: neomycin; TE: tetracycline; DX: doxycycline; MI: minocycline; E: erythromycin; MD: medemcyin; NOR: norfloxacin; OFX: ofloxacin; CIP: ciprofloxacin; VA: vancomycin; PB: polymyxin B; CC: clindamycin; SXT: trimethoprim-sulfamethoxazole; FZ: furazolidone; C: chloramphenicol.