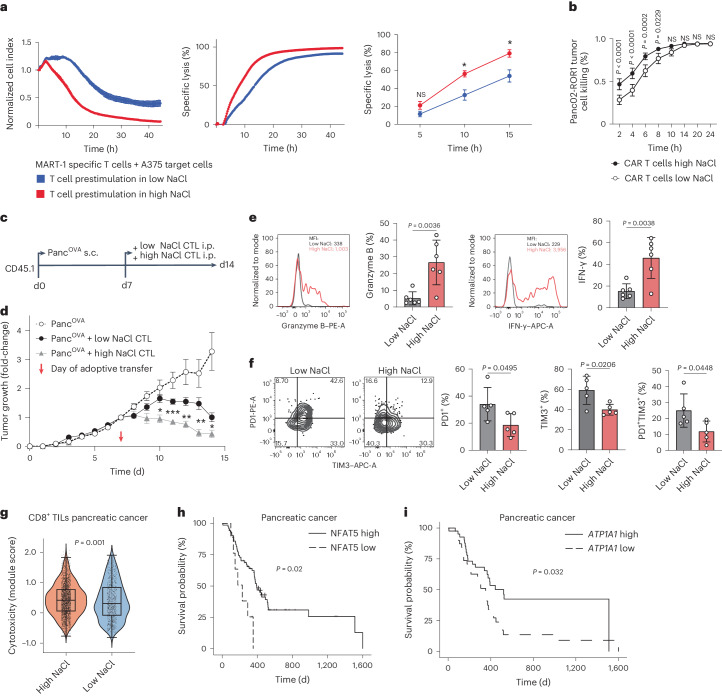
Fig. 6. NaCl licenses CD8+ T cells for killing of tumor cells in vitro and in vivo.
a, Real-time killing assay with nucleofected MART-1-specific T cells and A375 melanoma cell target cells at a 1:1 ratio under high and low NaCl conditions using the xCELLigence technology. Left, the normalized cell index; middle, the specific lysis; right, the cumulative quantification of 3T cell donors (n = 3 experiments; mean ± s.e.m.; two-way ANOVA, *P < 0.05). b, Murine ROR1 CAR T cells generated and cultured for 48 h under high and low NaCl conditions and then cocultured with ROR1-expressing target cells at a 10:1 ratio. Antigen-specific lysis of Panc02-ROR1 cells by CD8+ CAR T cells was determined at different time points (n = 3 independent experiments; mean ± s.d., two-way ANOVA). c, Experimental design. d, The tumor growth curves of subcutaneous tumors. Tumor growth was normalized to the tumor size on the day of CD8+ T cell injection (n = 7 (PancOVA), n = 6 (PancOVA + low NaCl control (CTL)), n = 6 (PancOVA + high NaCl CTL); mean ± s.e.m. two-way ANOVA with Tukey’s honestly significant difference (HSD), multiple-comparison test). e,f, Flow cytometric analysis of intratumoral CD45.2+CD8+ T cells 72 h after T cell transfer (n = 6 (e), n = 5 (f); mean ± s.d., two-tailed, unpaired Student’s t-test). g, ScRNA-seq and module score calculation for T cell cytotoxicity genes obtained from published reports55,56, validated with genes from GO:0001916 (P = 0.01). Intratumoral CD8+ T cells are shown from 56 patients with pancreatic cancer (from accession nos. GSE155698, GSE111672, GSE154778, GSM4293555 and PRJCA001063)40, integration of all cells: 10.5281/zenodo.6024273. CD8+ T cells were categorized into cells with a high and low NaCl signature based on the NaCl signature obtained from scRNA-seq of CD8+CD45RA– T cells treated under high versus low NaCl concentrations (top 60 upregulated DEGs; Supplementary Table 2; cutoff defined as module score ≥0 and <0 for high versus low NaCl signature, respectively; Wilcoxon’s rank-sum test). h,i, Kaplan–Meier tumor-free survival probability of patients from TCGA database diagnosed with pancreatic cancer. Patients were subgrouped by computing an optimal cutoff for NFAT5 (h) and ATP1A1 (i) expression. TPM values were normalized toward overall survival outcome. Number of patient samples: pancreatic cancer: n = 72 for NFAT5 high, n = 9 for NFAT5 low; n = 41 for ATP1A1 high; n = 40 for ATP2A2 low; significance of survival differences was determined using the Peto–Peto algorithm with the surv_pvalue function (method = ‘S1’) as implemented in the R package survminer.