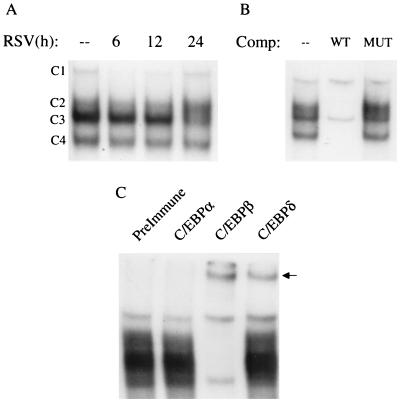
FIG. 10.
EMSA of RANTES NF-IL6 binding complexes in response to RSV infection. (A) Autoradiogram of time course. Nuclear extracts were prepared from control and RSV-infected cells at the indicated times and used for EMSA. Time following RSV infection is shown at the top. Four complexes (C1, C2, C3, and C4) are formed similarly in control and RSV-infected cells. (B) Competition (Comp) analysis. Nuclear extracts from A549 cells uninfected or infected for 12 h were used to bind to the NF-IL6 probe in the absence (–) or presence of 2 pmol of unlabeled WT or MUT competitor. C2, C3, and C4 are competed by the unlabeled oligonucleotide, indicating binding specificity. (C) Supershift assay. Nuclear extracts of A549 cells infected for 12 h were used in the EMSA in the presence of preimmune serum, anti-C/EBPα, anti-C/EBPβ, and anti-C/EBPδ. The addition of anti-C/EBPβ produces the disappearance of the specific complexes and the appearance of a supershifted band; the addition of anti-C/EBPδ also induces the appearance of a supershifted band.