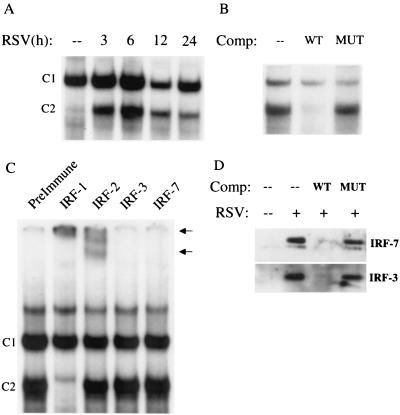
FIG. 6.
EMSA of RANTES ISRE binding complexes in response to RSV infection. (A) Autoradiogram of time course. Nuclear extracts were prepared from control and RSV-infected cells at the indicated times and used for EMSA. Time following infection is shown at the top. A single nucleoprotein complex 1 (C1) is formed in control cells, while a second complex (C2) appears following RSV infection. (B) Competition (Comp) analysis. Nuclear extracts from A549 cells infected for 12 h were used to bind to the ISRE probe; 2 pM unlabeled wild-type (WT) or mutated (MUT) competitor was included in the binding reaction, as indicated at the top. C1 is competed by the wild-type oligonucleotide but not by the mutant one, indicating binding specificity. (C) Supershift assay. Nuclear extracts of A549 cells infected for 12 h were used in the EMSA in the presence of preimmune serum and anti-IRF-1, -IRF-2, -IRF-3, and -IRF-7 antibodies. The anti-IRF-1 antibody induced the complete disappearance of C2 and the appearance of a supershifted band, which was also induced by the anti-IRF-2 antibody (as indicated by the arrows). (D) Microaffinity isolation-Western blot analysis for IRF-3 and -7 Control and 12-h-infected A549 nuclear extracts were affinity purified using biotinylated ISRE in the absence or presence of nonbiotinylated wild-type (WT) or mutated (MUT) competitor. After capture with streptavidin-agarose beads, complexes were eluted and assayed for IRF-3 and IRF-7 by Western blotting. IRF-3 and -7 are not present in the ISRE complex formed by control nuclear extracts, but they are strongly induced to bind to the ISRE following RSV infection. Binding is competed by the wild-type nonbiotinylated oligonucleotide but not the mutated one, indicating sequence specificity.