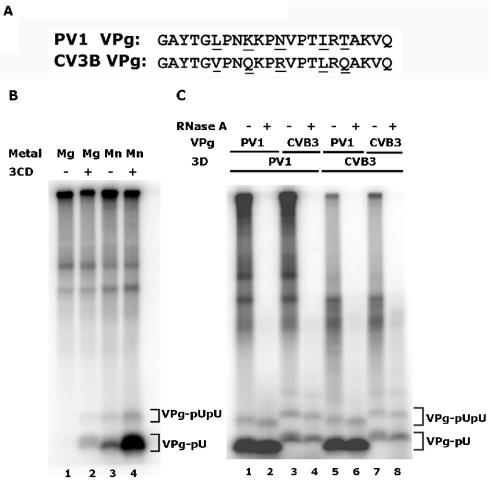
FIG. 1.
VPg uridylylation catalyzed by poliovirus (PV-1) or coxsackievirus B3 (CVB3) 3D polymerase using a poliovirus 2C RNA template in the presence of poliovirus 3CD. (A) The sequences of PV-1 and CVB3 VPg proteins are shown. Differences in sequence are underlined. (B) PV-1 VPg uridylylation by PV-1 3D polymerase using a 2C RNA template in the presence of magnesium acetate (lanes 1 and 2) or manganese chloride (lanes 3 and 4). Poliovirus 3CD was absent in lanes 1 and 3 and present in lanes 2 and 4. (C) VPg uridylylation by PV 3D polymerase (lanes 1 to 4) and CVB3 3D polymerase (lanes 5 to 8) using a 2C RNA template; all reactions contained PV 3CD. The identity of the VPg substrate, from PV or CVB3, is indicated. Products were digested with RNase A (100 μg/ml, 30°C, 60 min) in even-numbered lanes as indicated. Products were resolved in 12% polyacrylamide-Tris-Tricine gels. The positions of migration of VPg-pUpU and VPg-pU are indicated in both (B) and (C). The identity of VPg-pU and VPg-pUpU were confirmed by RNase and snake venom phosphodiesterase treatment; slightly greater differences in mobility are seen in this gel than in subsequent separations of VPgpU and VPgpUpU.