Fig. 1.
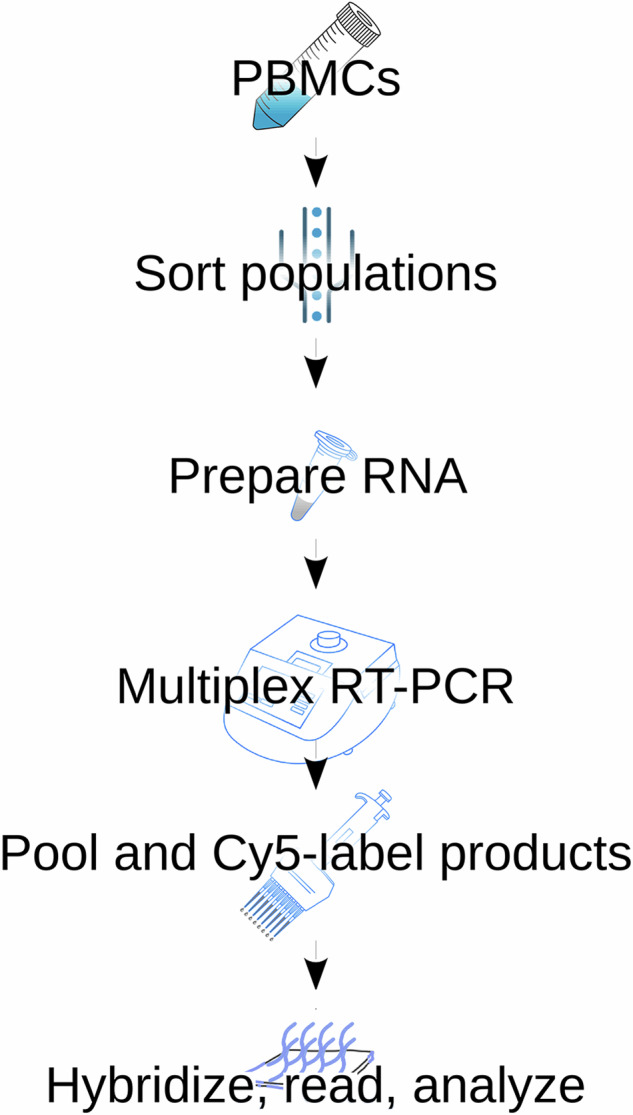
Experimental workflow. PBMCs were collected from healthy donors and sorted into different cell populations. RNA was prepared and its quality and concentration assayed according to standardized techniques. A total of 100 ng per reaction were used as input material to amplify GPCR transcripts in eight separate reactions each containing 50 GPCR specific primer pairs and aminoallyl-dUTP. Separate reactions with 10 ng of input material were used to determine those receptors expressed at higher levels. PCR reactions were pooled and Cy5 labelled on the amino-modified nucleotides. Labelled PCR products were hybridized to custom GPCR arrays that contained plus and minus sense oligonucleotides. Arrays were processed and scanned, and their signal extracted analyzed.
