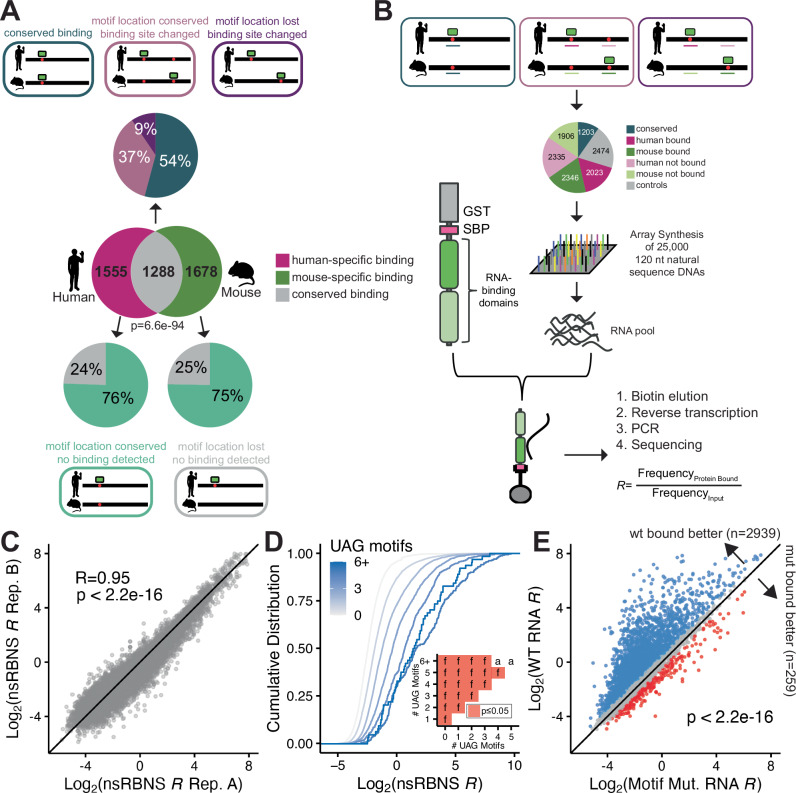
Fig. 1. Design and validation of natural sequence RNA bind-n-seq (nsRBNS).
A (Venn diagram) Transcript-level conservation of iCLIP UNK hits between human neuronal cells (SH-SY5Y) and mouse brain tissue. Significance determined via hypergeometric test. (Pie charts) Motif level conservation of iCLIP UNK hits between human neuronal cells (SH-SY5Y) and mouse brain tissue. B Design of natural sequence oligo pool and layout of nsRBNS. C Correlation plot of two experimental UNK nsRBNS replicates. Pearson’s correlation coefficient and p val included. D Cumulative distribution function of log2 nsRBNS enrichment of all oligos separated by UAG motif content. Inset shows significance values for all comparisons via two-sided KS test and corrected for multiple comparisons via the BH procedure. Red denotes significant (p ≤ 0.05). Values are as follows: a (ns), f (p ≤ 0.0001). E Scatter plot of log2 nsRBNS enrichment of wild-type (Y-axis) versus motif mutant (X-axis) oligos. Log2 change in enrichment (wt-mut) was calculated for each sequence pair: >0.5 defined as bound better in wt (blue), <−0.5 defined as bound better in mut (red), 0 ± 0.5 defined as similar binding (grey). Significance determined via paired, one-sided Wilcoxon test.