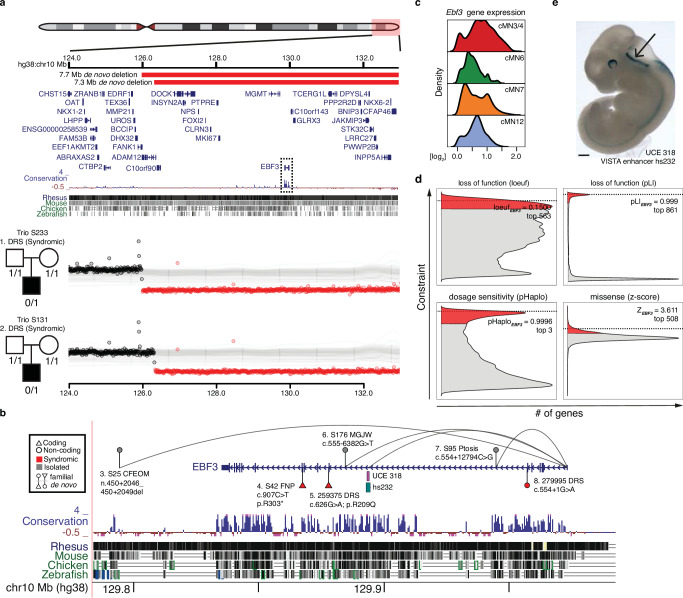
Fig. 5. An integrated coding/non-coding candidate allelic series for EBF3.
a Window depicting the terminal arm of chr10q (top). Large de novo deletions in two trios (middle, bottom) with simplex syndromic DRS (S233 and S131) overlap multiple coding genes including EBF3 (boxed), an exceptionally conserved gene at the coding and non-coding level. b Nominated coding and non-coding SNVs and indels connected to EBF3. For each variant, the subject’s WGS ID code, CCDD phenotype, and the variant coordinate in NG_030038.1 (and if coding or noncoding and if familial or de novo) is indicated. Variants 5 and 8 are reported previously in DECIPHER and elsewhere (10.1016/j.ajhg.2016.11.020). Peak-to-gene links containing variants connected to EBF3 are depicted by curved lines. EBF3 contains highly conserved non-coding intronic elements, including ultraconserved element UCE318 in intron 6, whose sequence drives strong expression in the embryonic hindbrain. c Imputed gene expression profiles for Ebf3. d EBF3 is exceptionally intolerant to loss-of-function, gene dosage, and missense variation. Density plots depict the genome-wide distribution of loss-of-function constraint (loeuf, pLI) (10.1038/s41586-020-2308-7; 10.1038/nature19057), probability of haploinsufficiency (pHaplo) (10.1016/j.cell.2022.06.036), and missense constraint (z-score) (10.1038/ng.3050). Respective scores exceeding thresholds of 0.35, 0.9, 0.84, and 2.0 are colored red. EBF3 (dotted lines) ranks as the 563rd, 861st, 3rd, and 508th most constrained gene in the genome, respectively. Distributions are rescaled for consistent signs and ease of visualization. e Lateral view of in vivo reporter assay testing UCE318 (VISTA enhancer hs232; n = 7 embryos), a putative EBF3 enhancer (peak-to-gene r = 0.42, FDR = 6.72 × 10−22). Strong reporter expression is observed in the embryonic hindbrain (arrow). Scale bar in e = 500 μm and is approximate based on E11.5 embryo average crown-rump length of 6 mm.