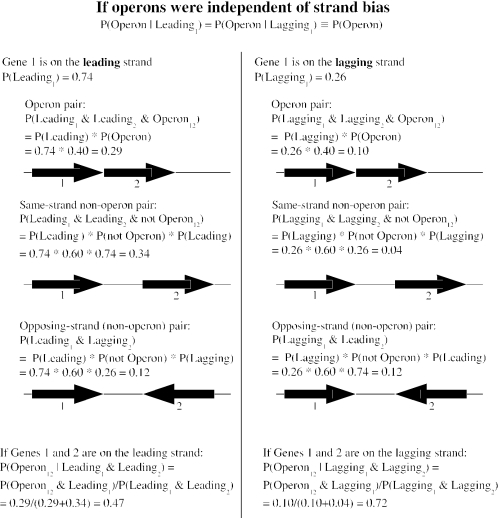
Figure 2.
If operons were independent of strand bias, then pairs of genes on the leading strand would be less likely to be in operons. We show the six probabilities for two adjacent genes to be in the same operon (or not) and on the same strand (or not), given that the first gene is on the leading strand (or not). The probabilities are derived from the null hypothesis that operons do not affect strand bias and from 74% of genes being on the leading strand of B.subtilis. The estimate is that 40% of genes are in an operon with their downstream gene from the output of our operon predictions (also for B.subtilis). The statistical effect arises from considering only same-strand pairs: if operons were independent of strand bias, then most lagging-strand pairs but only about half of leading-strand pairs would be in the same operon.