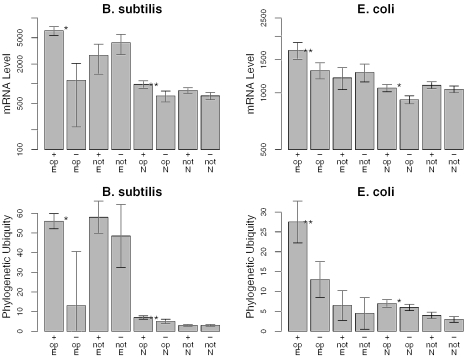
Figure 5.
After controlling for essentiality, genes in predicted operons are more highly expressed and more phylogenetically ubiquitous if they are on the leading strand. We show the median of expression level (top) and phylogenetic ubiquity (bottom) for essential (‘E’) or non-essential (‘N’) genes on the leading (‘+’) or lagging (‘−’) strand predicted to be in operons (‘op’) or not, from B.subtilis (left) and E.coli (right). For each group of genes, the gray bars show the median and the error bars indicate the 90% confidence interval of the median. Significant differences between genes on the leading and lagging strands are marked with ‘**’ (P < 0.005, Wilcoxon rank sum test) or ‘*’ (P < 0.05). As in Figure 3, non-essential genes in predicted operons with essential genes are not included.