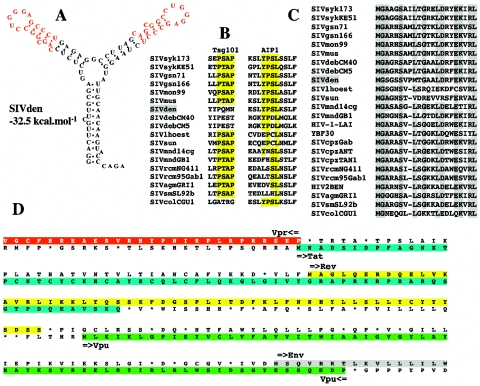
FIG. 6.
Features of SIVden. (A) Secondary structure for SIVden TAR with the lowest free energy value (−32.5 kcal/mol). Red letters indicate sequence conserved between the two stems. (B) Alignment of primate lentiviral Gag P6 protein sequences. Only the two putative binding sites for the Tsg101 [P(T/S)AP] and AIP1 [YP(D/S)L] proteins are shown. Conserved amino acids are highlighted in yellow. (C) Alignment of the N terminus of the Gag gene. Dashes indicate space introduced to optimize the alignment. Conserved amino acids are highlighted in gray. Dots indicate amino acid identity at a given residue. (D) Organization of the vpr-env region. The vpr and tat ORFs overlap on a single amino acid residue. The tat ORF is highlighted in blue, while the rev ORF is highlighted in yellow; the vpu ORF is highlighted in green, and the N terminus of the env ORF is highlighted in gray.