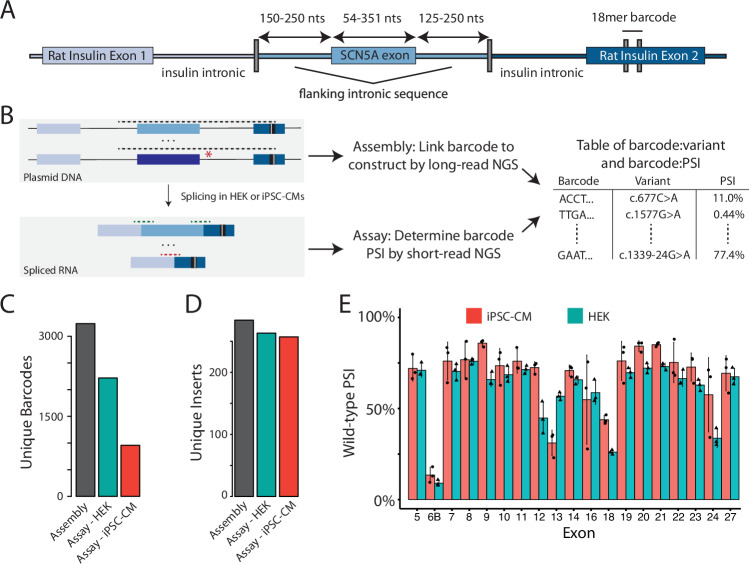
Fig. 2. ParSE-seq assay in HEK cells and iPSC-CMs.
A Detailed schematic of the ParSE-seq barcodable minigene plasmid. A previously used minigene vector was mutagenized to introduce a restriction site into the 3′ rat insulin exon 2. After digestion of a pool of minigene plasmids with SCN5A WT and variant inserts (middle), an 18-mer barcode was subcloned into the downstream exon (see Supplementary Fig. 2 for complete barcoding steps). Nts nucleotides. B Overview schematic of assembly and assay steps, and subsequent integration. Dashed lines represent amplicons for long- and short-read next generation sequencing (NGS). Following barcode insertion into the digested plasmid pool, a long-read PCR amplicon was used to link the barcode to the SCN5A insert (assembly). The pool was transfected into HEK cells and iPSC-CMs, after which short-read RNA-seq was used to link the barcode to splicing outcomes (assay; PSI percent spliced in). Assembly and assay data were merged by barcode for subsequent analysis steps (Supplementary Fig. 3). C Barcode counts for the assembly, and recovered barcodes present across three replicates in HEK and iPSC-CM assays. More barcodes were detected from the more easily transfected HEK cells than iPSC-CMs. Raw data available in Source Data and Supplementary Data 3. Black (library, pretransfection), red (iPSC-CMs), green (HEK). D Unique WT or variant inserts covered by barcodes in (C). Despite lower total barcode recovery in (C), most inserts are still recovered with the high stoichiometry of barcodes: inserts. Raw data available in Source Data and Supplementary Data 3. E PSI for all WT exons in iPSC-CMs and HEK cells. Data are averaged across three replicates and error bars represent the standard error of the mean. Red indicates iPSC-CMs, green indicates HEK cells. Raw data available in Source Data.