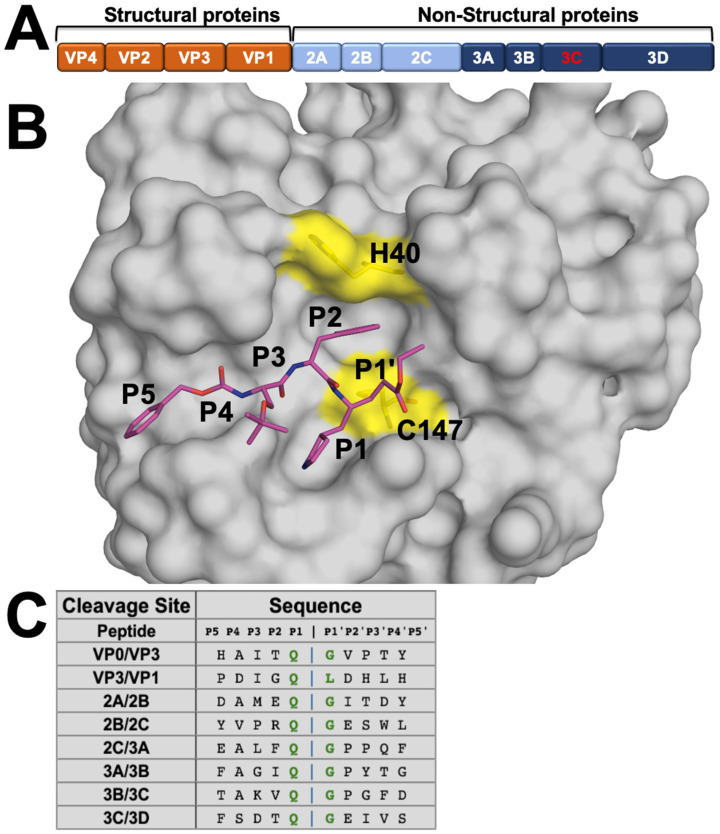
Figure 1.
EV68-3C protease substrates and structure. (A) EV68 polyprotein prior to cleavage by the 3C protease into structural and non-structural viral proteins. (B) Crystal structure of EV68-3C protease in surface representation bound to a peptidomimetic inhibitor (SG-85, in magenta sticks) with cysteine protease’s catalytic dyad residues (H40 and C147) highlighted in yellow (PDB ID: 3ZVF). The inhibitor is labeled with corresponding P5 to P1′ moieties. (C) Amino acid sequences of EV68-3C protease cleavage sites in the viral polyprotein, with a conserved glutamine (Q) in the P1 position, generally followed by a glycine (G) at P1′. The sequences are highly diverse distal to the cut site (denoted by the blue vertical bar).