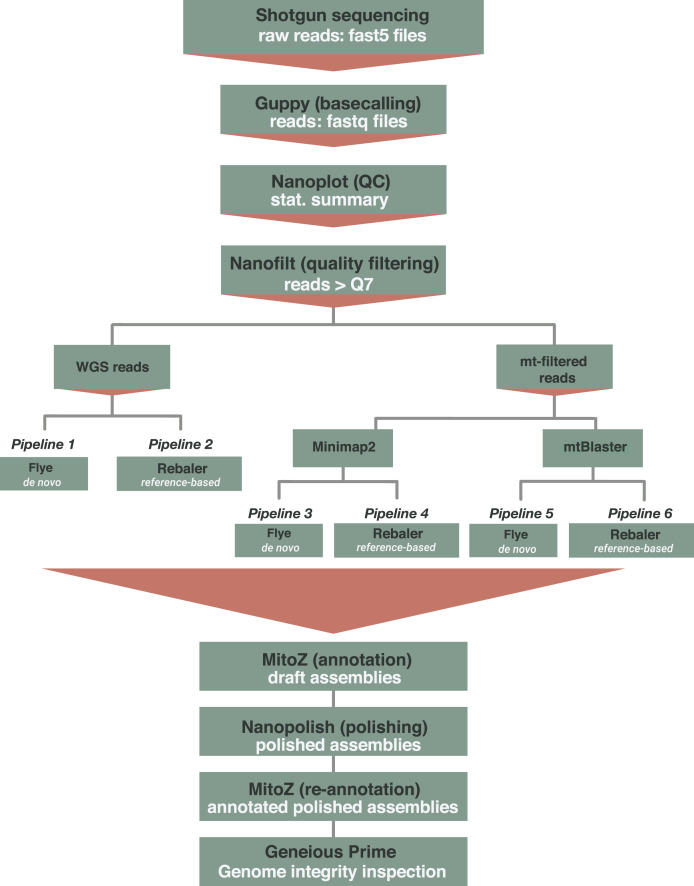
Figure 2. Computational workflow implemented for the reconstruction of mitochondrial genomes from ONT shotgun sequencing data.
Pipelines 1 and 2 use WGS reads and use de novo (Flye) and reference based (Rebaler) assemblers. Pipelines 3 to 6, initially filter the mitochondrial reads implementing Minimap2 (pipelines 3 and 4) or mtBlastr (pipeline 5 and 6) to subsequently execute the assemble step (Flye and Rebaler). The final steps (below red triangle) that include annotation and polishing are common to all six pipelines.