FIGURE 3.
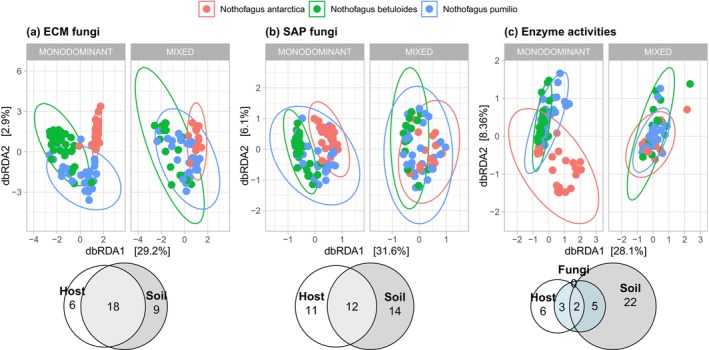
Distance‐based redundancy analysis of (a) ectomycorrhizal (ECM) and (b) saprotrophic (SAP) fungal community composition based on Bray‐Curtis distances and stand (monodominant or mixed) as a condition: Soil pH (F = 49.42***), host (F = 6.66***), soil moisture (F = 4.36***) and available N (F = 2.16**) were the best predictors of ECM fungal community structure, and 18% of the variation was explained by the interaction of host and soil variables, regardless of stand. Host (F = 22.88***), soil pH (F = 13.96***), and soil moisture (F = 10.93***) were the best predictors of SAP fungal community structure, and 12% of the variation was explained by the interaction of host and soil variables. Differences in fungal community composition were non‐significant between co‐occurring hosts in mixed forests. (c) Redundancy analysis of soil enzyme activities based on Euclidean distances, with samples colored by Nothofagus species: Soil moisture (39.62***), soil pH (F = 13.23***), host (F = 10.96***), and available P (F = 7.37***) were the best predictors of the variation in enzyme activities, with soil variables explaining 22% of the variation in enzyme activities.
