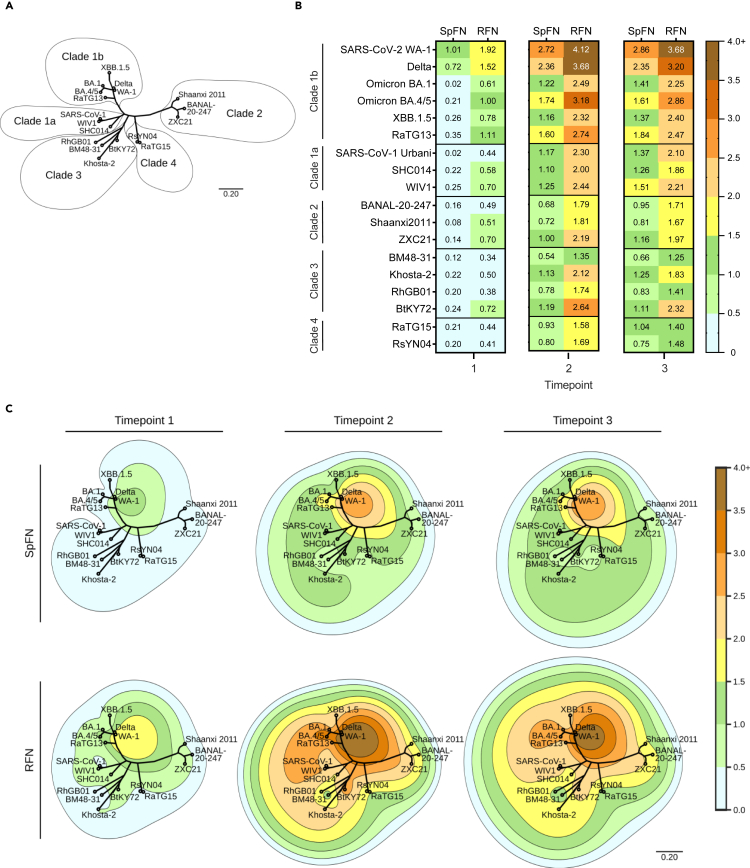
Figure 3.
Cross-reactive RBD-binding response against genetically distant sarbecoviruses
(A) Phylogenetic tree, based on the RBD amino acid sequences of SARS-CoV-2 VoC and sarbecoviruses.
(B) Average serum binding responses (nm) to sarbecovirus RBD molecules measured by BLI for SpFN- and RFN-immunized animals (timepoints indicated at base of the plot).
(C) Contour-phylogenetic plots of binding of SpFN and RFN immunized animal sera to sarbecovirus RBD molecules. X and Y coordinates are determined by a phylogenetic tree of the RBD amino acid sequences, and contour height determined by detected BLI serum binding. Interpolation and extrapolation of binding data (elevation) between points on the phylogenetic tree was determined by a thin plate radial basis function (r2∗log(r)) using the scipy python package. For clarity, extrapolated values higher than the maximum measured binding were truncated at the maximum binding for the timepoint.