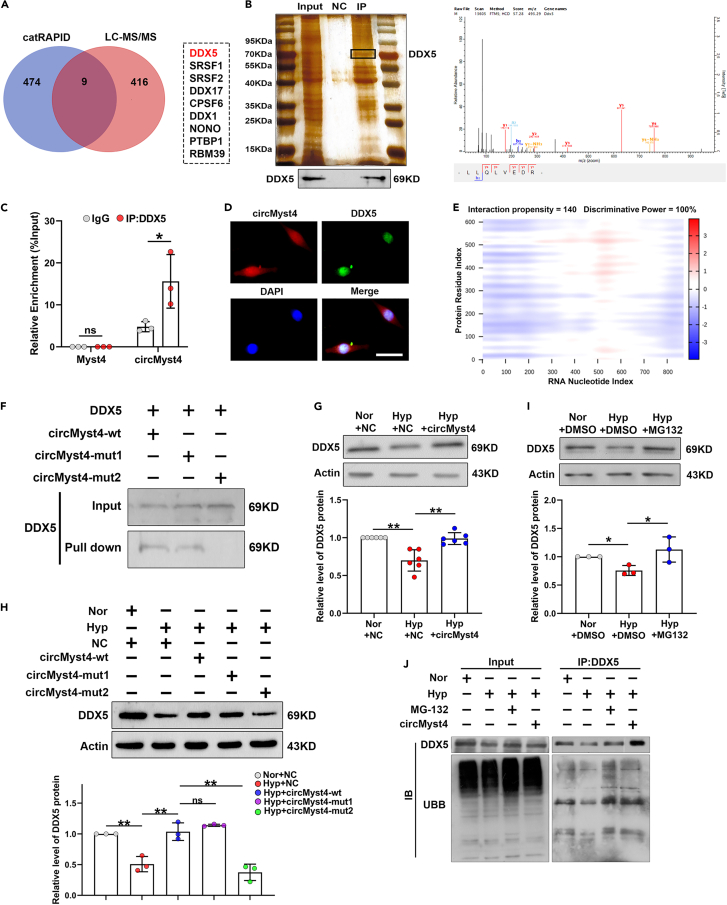
Figure 4.
The association of circMyst4 with DDX5
(A) catRAPID databases and Mass spectrometry analysis of the binding protein of circMyst4.
(B) RNA pull-down and western blotting analysis were used to identify proteins associated with circMyst4 (left), and mass spectrometry of specific segments of DDX5 (right).
(C) RIP-PCR analysis was performed to prove the association of circMyst4 to DDX5 protein (n = 3).
(D) The colocalization of circMyst4 and DDX5 in PASMCs. Scale bars, 100 μm (n = 3). Green color denotes DDX5, stained with FITC, and blue color denotes nucleus, stained with DAPI, whereas circMyst4 probes were labeled with Cy3 (red).
(E) Predicted interaction of circMyst4 (nucleotide positions 326-377nt and 701-752nt) and DDX5 protein.
(F) RNA pull-down and western blotting analysis were used to identify the binding site of DDX5 to circMyst4 (mut1, circMyst4 nucleotide positions 326-377nt; mut2, circMyst4 nucleotide positions 701-752nt).
(G and H) Detection of the level of DDX5 protein in PASMCs treated with overexpressing circMyst4 wild type (circMyst4-wt) or circMyst4 mutant type (circMyst4-mut) (n = 6, 3).
(I) Western blotting analysis of DDX5 in PASMCs treated with MG-132 (an inhibitor of ubiquitin protease system) (n = 3).
(J) Co-immunoprecipitation (CoIP) analysis was performed using anti-DDX5, followed by probing with anti-UBB in PASMCs transfected with circMyst4 overexpressing plasmid or treated with MG-132 (n = 3). Data are shown as means ± SD. Statistical analysis of the graph (C) was performed with Student’s t test. Statistical analysis of the graph (G, H, and I) was performed with one-way ANOVA followed by Dunn’s post-test. Hyp, hypoxia; Nor, normoxia; ns, not significantly different. ∗p < 0.05, ∗∗p < 0.01.