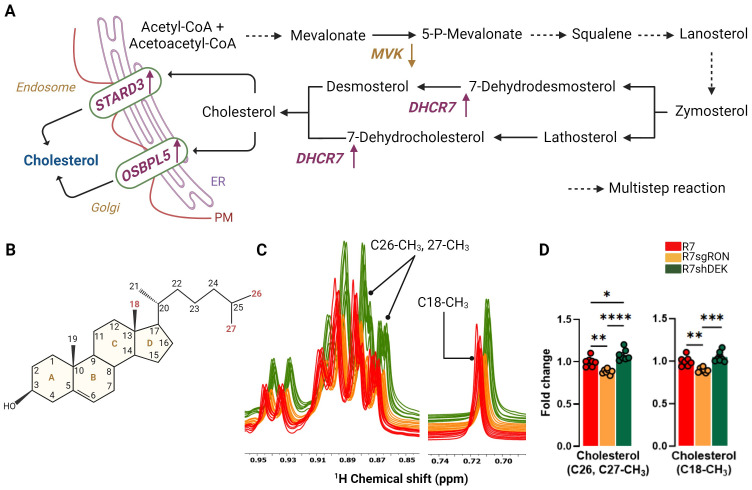
Figure 2.
RON stimulates cholesterol metabolism. (A) Cholesterol biosynthesis pathway. Multistep reactions are illustrated with discontinuous arrows. Relevant enzymes from the KMplot.com-derived lipid metabolism gene signature are shown where appropriate, with an upward arrow indicating that gene induction is associated with worse patient outcomes and a downward arrow indicating that gene suppression is associated with worse patient outcomes. (B) Chemical structure of cholesterol. The carbon positions highlighted in red represent the cholesterol atoms quantified by NMR and compared between cell lines. (C) Comparison of 1D 1H-NOESY spectra of the lipid fraction from R7, R7sgRON, and R7shDEK corresponding with the cholesterol peaks analyzed by 1H-NMR. (D) Intracellular fold change of cholesterol measured from C26, C27-CH3, and C18-CH3 carbon positions and quantified by 1H-NMR and expressed as a fold change of the control group (R7). Bar plots indicate mean ± SEM. Replicates of each cell line are shown as individual dots (n = 7). Statistical significance was assessed using one-way ANOVA. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. PM, plasma membrane; MVK, mevalonate kinase; DHCR7, 7-dehydrocholesterol reductase; STARD3, StAR-related lipid transfer domain containing 3; OSBPL5, oxysterol binding protein like 5.