Fig. 5: Introgressed SAVs present in modern humans were shared across archaic individuals and are associated with increased tissue specificity.
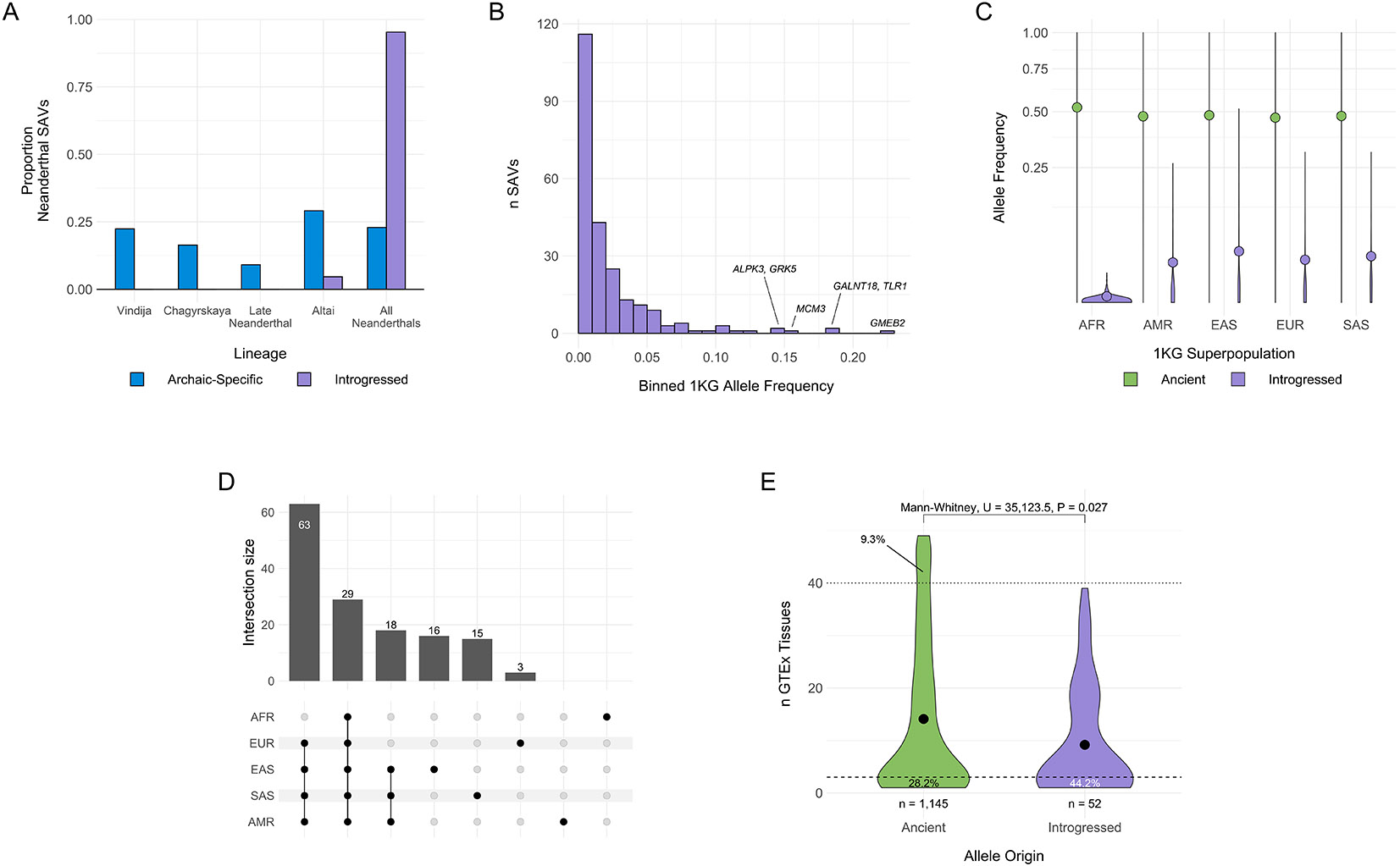
(A) Histograms comparing distributions of the presence of all Neanderthal SAVs (blue) and introgressed SAVs (purple) in different sets of Neanderthal individuals. Introgressed SAVs are significantly older than expected from all Neanderthal SAVs. We focused on Neanderthal lineages because of low power to detect introgressed Denisovan SAVs. All data are presented in Supplementary Table 10. (B) Allele frequency distribution for introgressed SAVs per [38]. Allele frequencies represent the mean from the 1KG African, American, East Asian, European, and South Asian superpopulation frequencies. (C) Allele frequency distributions for SAVs present in both archaic and modern individuals stratified by 1KG superpopulation and origin (ancient vs. introgressed) per [38]. Ancient SAVs (n = 2,252) are colored green and displayed on the left, while introgressed SAVs (n = 237) are colored purple and displayed on the right per superpopulation. AFR = African, AMR = American, EAS = East Asian, EUR = European, SAS = South Asian. The colored dot represents the mean allele frequency for each set. The y-axis is square root transformed. (D) The number of introgressed SAVs with a minimum allele frequency of at least 0.01 in each modern human population. We display all individual populations, the non- African set, and Asian/American set here. See Supplementary Fig. 15 for all sets. In all panels introgressed SAVs and frequencies are as defined by [38]. (E) The distribution of the number of GTEx tissues in which an ancient or introgressed SAV (per [38]) was identified as an sQTL. Introgressed variants are significantly more tissue-specific. We defined “tissue-specific” variants as those occurring in 1 or 2 tissues and “core” sQTLs as those occurring in > 40 of the 49 tissues. The dashed and dotted lines represent these definitions, respectively. The proportion of SAVs below and above these thresholds are annotated for each allele origin.
