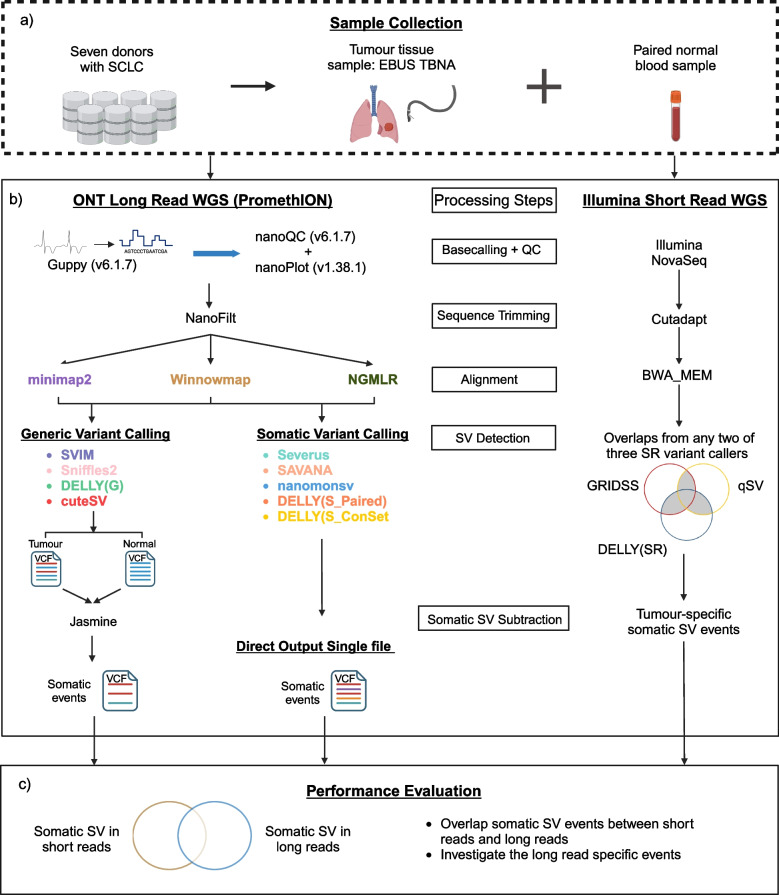
Fig. 1.
Overview of study design to benchmark alignment and somatic SV calling. The workflow includes multiple steps. a Sample collection. DNA was extracted from seven tumour lung samples collected from EBUS-TBNA and seven matching blood samples. b The normal and tumour DNA underwent whole genome sequencing (WGS) using ONT PromethION for long-read sequencing (LRS) and Illumina NovaSeq for short-read sequencing (SRS). The processing steps for identifying somatic SV events in both long-read and short-read data used different tools but followed a similar process: sequence base calling and alignment, followed by the application of various variant calling methods for SV detection. In LRS, three aligners (minimap2, Winnowmap, and NGMLR) were evaluated, in combination with two approaches (generic and somatic calling) for detecting somatic SVs. The four generic SV callers were cuteSV, DELLY (G), Sniffles2, and SVIM, which required manual subtraction to determine somatic SVs with Jasmine. The somatic callers were DELLY (S_Paired), DELLY (S_ConSet), nanomonsv, SAVANA, and Severus. c The performance of each approach in LRS was evaluated by comparing the somatic SVs identified to high-confidence somatic SV events (obtained from SRS of the same samples and called by two or more of these approaches: qSV, DELLY, and GRIDSS)