Abstract
Motivation
The microbes in human body play a crucial role in influencing the functions of drugs, as they can regulate the activities and toxicities of drugs. Most recent methods for predicting drug–microbe associations are based on graph learning. However, the relationships among multiple drugs and microbes are complex, diverse, and heterogeneous. Existing methods often fail to fully model the relationships. In addition, the attributes of drug–microbe pairs exhibit long-distance spatial correlations, which previous methods have not integrated effectively.
Results
We propose a new prediction method named DHDMP which is designed to encode the relationships among multiple drugs and microbes and integrate the attributes of various neighbor nodes along with the pairwise long-distance correlations. First, we construct a hypergraph with dynamic topology, where each hyperedge represents a specific relationship among multiple drug nodes and microbe nodes. Considering the heterogeneity of node attributes across different categories, we developed a node category-sensitive hypergraph convolution network to encode these diverse relationships. Second, we construct homogeneous graphs for drugs and microbes respectively, as well as drug–microbe heterogeneous graph, facilitating the integration of features from both homogeneous and heterogeneous neighbors of each target node. Third, we introduce a graph convolutional network with cross-graph feature propagation ability to transfer node features from homogeneous to heterogeneous graphs for enhanced neighbor feature representation learning. The propagation strategy aids in the deep fusion of features from both types of neighbors. Finally, we design spatial cross-attention to encode the attributes of drug–microbe pairs, revealing long-distance correlations among multiple pairwise attribute patches. The comprehensive comparison experiments showed our method outperformed state-of-the-art methods for drug–microbe association prediction. The ablation studies demonstrated the effectiveness of node category-sensitive hypergraph convolution network, graph convolutional network with cross-graph feature propagation, and spatial cross-attention. Case studies on three drugs further showed DHDMP’s potential application in discovering the reliable candidate microbes for the interested drugs.
Availability and implementation
Source codes and supplementary materials are available at https://github.com/pingxuan-hlju/DHDMP.
1 Introduction
The human body hosts a diverse community of microbes, including bacteria, viruses, protists, archaea, and fungi (Gill et al. 2006, De Lorenzi-Tognon et al. 2023). These microbial communities are closely linked to human health (Aggarwal et al. 2022, VanEvery et al. 2023), with studies suggesting that they can promote human metabolism and enhance the immune system (Ventura et al. 2009, Sommer and Bäckhed 2013). However, disruptions in these communities can lead to diseases such as hypertension and diabetes (Wen et al. 2008, Xiong et al. 2021).
Microbes can also chemically modify drugs to enhance their efficacy (Zimmermann et al. 2019, Kumbhare et al. 2023). For example, Vich Vila et al. (2020) have shown that the human gut microbiota is involved in drug metabolism processes, and compounds present in these drugs can promote the growth of gut bacteria. Conversely, microbes can develop resistance to drugs, negatively impacting human health (Uddin et al. 2021, Li et al. 2023). Therefore, predicting drug–microbe associations is beneficial for understanding drug mechanisms of action and drug–microbe interaction mechanisms.
Recently, the computational methods were proposed for predicting the miRNA–disease associations (Li et al. 2021), drug-target interactions (Li et al. 2017, Song et al. 2024). Computational prediction methods also have proven to be effective in predicting candidate microbes associated with drugs and screening reliable candidates for subsequent biological experiments. Researchers have proposed several approaches for predicting drug–microbe associations. Methods such as GCNMDA (Long et al. 2020a), EGATMDA (Long et al. 2020b), and SCSMDA (Tian et al. 2023) use simple graph convolutional networks (Kipf and Welling 2017) to aggregate neighbor information without differentiating between neighbor types. GSAMDA (Tan et al. 2022) and MDASAE (Fan et al. 2023) use sparse autoencoders, while Graph2MDA (Deng et al. 2022) employs a variational graph autoencoder (Kipf and Welling 2016) to learn node representation in the drug–microbe heterogeneous graph. However, these methods do not capture the complex relationships among multiple drugs and microbes. GACNNMDA (Ma et al. 2023) uses a convolutional neural network to learn features of drug–microbe node pairs but ignores the spatial dependencies between attributes of node pairs.
We present DHDMP, a method driven by node category-sensitive hypergraph convolutional networks, graph convolutional network with cross-graph feature propagation (GCNFP), and spatial cross-attention (SCA) for predicting drug-related microbes. Our method fuses the characteristics of multiple drugs and microbes that are related, and integrates the heterogeneity of multiple types of nodes and the long-distance spatial correlations of drug–microbe node pairs. The contributions of this article are summarized as follows:
A microbe might affect the functions of multiple drugs, and multiple microbes are usually involved into the process that a drug exert its function. Thus, there are complex relationships among multiple drugs and multiple microbes. The hyperedges are designed to denote these relationships from multiple perspectives, and their embeddings are learnable. The learnable strategy is helpful for forming the hypergraph topology with dynamic changes.
Most previous methods based on hypergraph learning only integrated the hypergraph topology and the node attributes and they ignored the heterogeneity of feature distributions of multiple categories of nodes. We designed their category features for each category of nodes to discriminate the heterogeneity of drug and microbe nodes. A node category-sensitive hypergraph convolutional networks (NHCN) is presented to encode the dynamic hypergraph topology and the node heterogeneity.
A homogeneous graph composed of drug nodes, a homogeneous graph composed of microbe node, and a heterogeneous graph composed of both drug and microbe nodes, are established to reflect the similarities among the nodes from the same category and the associations among the nodes from different categories. In these graphs, a target node has its homogeneous neighbors and heterogeneous neighbors which reveal its characteristics from diverse perspectives. A feature propagation strategy is also proposed to supplement the node features from homogeneous graph to the heterogeneous graph learning. A GCNFP was designed to fuse the attributes of these neighbors and their topologies.
The attribute embedding of a pair of drug and microbe node was constructed and there are long-distance spatial correlations among some attributes. We proposed a SCA to separate the pairwise attributes into multiple patches and integrate the long-distance correlations among these patches. The experimental results showed that our method achieved higher prediction performance than the compared advanced methods.
2 Materials and methods
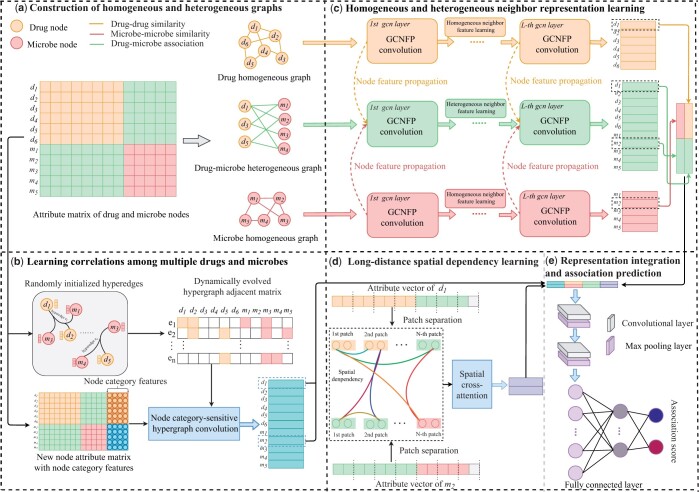
We propose a new prediction model, DHDMP (as shown in Fig. 1), designed to predict the microbes associated with specific drugs. DHDMP consists of three components that respectively learn the complex relationships among multiple drugs and microbes, information from homogeneous and heterogeneous neighbors, and long-distance spatial correlations between pairs of drug and microbe nodes. We construct a hypergraph to represent the associations among multiple drugs and microbes and design a NHCN to capture the complex relationships among drugs and microbes. To integrate features from both homogeneous and heterogeneous neighbors of the target drug or microbe node, we propose a GCNFP. We introduce a SCA mechanism to capture long-distance spatial correlations between the features of drug and microbe node pairs. These components work together to enhance the predictive power of DHDMP in identifying drug–microbe associations.
Figure 1.
Overall framework of DHDMP model. (a) Construct the drug and microbe homogeneous graphs and drug–microbe heterogeneous graph. (b) Learn complex relationships among multiple drugs and microbes by NHCN. (c) Learn homogeneous and heterogeneous neighbor representations based on GCNFP. (d) Encode long-distance spatial correlations by SCA. (e) Integrate multiple representations and estimate the association score
2.1 Dataset
The associations between drugs and microbes, drug chemical structure similarity, Gaussian kernel similarity, and microbe sequence similarity were obtained from previous research (Deng et al. 2022). The dataset contains associations on 2470 pairs of drugs and microbes, covering 1373 drugs and 173 microbes. The data on drug–microbe associations were originally obtained from the Microbe–Drug Associations Database (MDAD) (Sun et al. 2018). Furthermore, 365 984 drug interactions were obtained from the DrugBank database (Wishart et al. 2018), while the raw microbe gene sequence data were retrieved from the National Center for Biotechnology Information (NCBI) database.
2.2 Construction of homogeneous and heterogeneous graph for drugs and microbes
We construct a drug homogeneous graph and a microbe homogeneous graph . Here, and represent the sets of drug and microbe nodes, respectively. An edge connects drug nodes , while microbe nodes are connected by . Additionally, we construct a drug–microbe heterogeneous graph , where an edge connects a drug node and a microbe node .
The drug similarity matrix and microbial similarity matrix are defined as follows,
| (1) |
| (2) |
where and represent the number of drugs and microbes, respectively. For a pair of drug nodes and , their structural similarity is calculated by the SIMCOMP2 (Hattori et al. 2010) measurement based on their chemical structure graphs. The Gaussian kernel similarity between the i-th drug and the j-th one is computed based on the two groups of microbes interacted with them,
| (3) |
where is the ith row of the drug–microbe association matrix. represents the normalized kernel bandwidth,
| (4) |
where is the original bandwidth and it was set to 1. The final drug similarity is obtained by averaging the structural similarity and the Gaussian kernel similarity. Principal component analysis (PCA) (Weber et al. 2016) is used to calculate the similarity between microbe nodes and based on their gene sequences. These calculation methods were proposed by Long et al. (2020a). Since each row of the drug similarity matrix and the microbe similarity matrix contains the associations of the respective drug or microbe with all drugs and microbes, we can regard these matrices as the adjacency matrices of the drug homogeneous graph and the microbe homogeneous graph, respectively, and rename them as and .
The adjacency matrix of the drug–microbe heterogeneous graph is defined as,
| (5) |
where denotes the known association between the drug node and the microbe node , and otherwise. The node attribute embedding matrix includes the drug similarity matrix , the microbe similarity matrix , and the drug–microbe association matrix ,
| (6) |
where represents the transpose matrix of . If a pair of microbes and drugs are associated or similar with more common drugs and microbes, they are more likely to be associated. The th row of matrix , denoted as , contains the similarities and associations between node and all drugs and microbes. Therefore, can be considered the attribute of the node.
2.3 Multiple drugs and microbes association learning based on NHCN
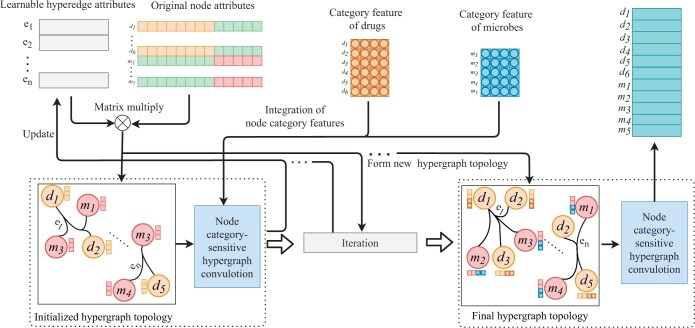
A microbe often participates in multiple drug function processes, and multiple microbes may simultaneously participate in the same drug function process, leading to complex relationships between multiple drugs and microbes. To capture such relationships, we establish multiple hyperedges and form a drug–microbe hypergraph. We construct a hyperedge embedding matrix , where is randomly initialized and trainable during the training process, and is the number of hyperedges. indicates the tendency of the -th node to belong to the -th hyperedge. Combining the node attribute embedding matrix , we form a hypergraph with dynamically changing topology, where its adjacency matrix is denoted as ,
| (7) |
The hypergraph topology is continuously updated during the training process, which is helpful for better reflecting the complex relationships among multiple drugs and microbes.
In the hypergraph, there are two types of nodes: drugs and microbes, and their feature distribution is heterogeneous. We design category features to represent node types. The category feature matrix for all drug nodes is denoted as , and for microbe nodes, it is denoted as , where represents the dimension of the node category features. are randomly initialized, and the category features of nodes with the same type have the same initial vector. are stacked vertically to form the category feature matrix for all drug and microbe nodes. The node attribute matrix embedded with category features is referred to as . To encode the complex relationships among multiple drugs and microbes, we design NHCN (node category-sensitive hypergraph convolution network). After layer encoding, we obtain the feature map ,
| (8) |
where is the activation function, is the number of convolution layers of the hypergraph. and are the degree matrices of nodes and hyperedges, corresponding respectively to the sum of rows and columns in the hypergraph adjacency matrix. is an attribute matrix embedded with category features. We perform hypergraph convolution on all nodes and then update the hyperedge embedding matrix. The updated hyperedge embedding matrix is multiplied with the node attributes to form a new hypergraph topology. We perform hypergraph convolution on the new hypergraph topology and repeat the iteration process (as shown in Fig. 2). After rounds of hypergraph convolution, we obtain multi-node association representations and of and , respectively. and are stacked to form the multi-node association representation for drug and microbe node pairs,
| (9) |
Figure 2.
Illustration of forming and updating the dynamic hypergraph topology
2.4 Homogeneous and heterogeneous neighbor representation learning based on GCNFP
Neighboring nodes of a target node can reflect the characteristics of that node, we aggregate the features of neighboring nodes to form new features for the target node. A target node has both neighbors of the same type (homogeneous neighbors) and neighbors of different type (heterogeneous neighbors), each offering unique perspectives on the characteristics of the target node. The homogeneous neighbors of the target drug (or microbe) are nodes that have similarities, which from drug (or microbe) homogeneous graph. The heterogeneous neighbors associated with the target drug (or microbe) come from the drug–microbe heterogeneous graph, which reflects relationships between nodes of different types. Thus, the homogeneous neighbors reflect the characteristics of the target node from the perspective of similarity, while the heterogeneous neighbors reveal the characteristics of the target node from the perspective of association. To capture the distinct features reflected by homogeneous and heterogeneous neighbors of the target node, we design the GCNFP. First, we normalize the adjacency matrix of the drug–microbe heterogeneous graph to obtain (Kipf and Welling 2017),
| (10) |
here, represents the Laplacian normalization of , where is the diagonal matrix and represents the number of non-zero entries in the row vector of the adjacency matrix . Similarly, we compute normalized adjacency matrices and for the microbe and drug homogeneous graphs, respectively. We define the drug–microbe homogeneous adjacency matrix and the drug–microbe heterogeneous adjacency matrix as follows,
| (11) |
where is a matrix with all elements being 0. After the -th graph convolutional encoding layer, we can obtain the -th order homogeneous neighbor representations for all drugs and microbes,
| (12) |
where represents the initial node attribute matrix, denotes the parameter matrix, and is the total number of layers in the GCNFP. Following graph convolutional iterations, we can derive first to -th order homogeneous neighbor representations for both drugs and microbes. Given that these representations from homogeneous neighbors contain similarity information of drugs and microbes, while those from heterogeneous neighbors only encode association information between drugs and microbes, we devised a node feature propagation strategy (NFPS). This strategy leverages the information from homogeneous neighbors as auxiliary data to enhance the learning process of features from heterogeneous neighbors. Consequently, the -th layer of heterogeneous neighbor representations is expressed as,
| (13) |
where denotes the concatenation operation, and signifies the parameter matrix of the GCNFP. As each drug (microbe) node has its own unique information, we set a learnable attention coefficient for each node to capture the unique information. The attention coefficient vector of all the nodes is and it is randomly initialized. The operation may convert the vector into a diagonal matrix . denotes the node attribute matrix of th layer, containing detailed information about each node. To construct a comprehensive homogeneous neighbor representation for and , we concatenate the homogeneous neighbor representations from the -th to the -th order. This yields multi-order homogeneous neighbor representations denoted as ,
| (14) |
We utilize a fully connected layer to transform the high-dimensional homogeneous neighbor representations , which contain multi-order neighbor information, into a low-dimensional feature space. This transformation yields as follows,
| (15) |
where and represent learnable parameter matrices, while and are bias vectors. Similar to formulas (14) and (15), we derive heterogeneous neighbor representations and for nodes and . We combine these representations with the homogeneous neighbor representations and to obtain the homogeneous and heterogeneous representations of nodes and , denoted as ,
| (16) |
2.5 Long-distance spatial representation learning based on SCA
If and are similar or associated with more the same drugs and microbes, their attribute vectors exhibit more similarity in feature distribution. We reshape the attribute vectors of into , where represents patch size and . If is not divisible by , we pad the end of and with zeros until is divisible by . The reshaped matrices and are divided into patches, where patches with similar features might be widely dispersed in space. The partitioning of attribute patches is helpful for designing an SCA mechanism to capture the spatial correlations among the patches (as shown in Fig. 3).
Figure 3.
Encoding the long-distance correlations among multiple attribute patches of a pair of drug and microbe nodes
We introduce a multi-head attention mechanism (Dosovitskiy et al. 2020) to calculate attention from different perspectives to reduce bias in the learning process. Taking the -th head also the last head attention as an example, we employ distinct linear transformations to produce drug query matrices , key matrices , and value matrices for each patch. These transformations are defined as follows,
| (17) |
where, are weight matrices. For all the patches of a pair of drug and microbe nodes, , , are the shared weight matrices. It is beneficial for jointly capturing the spatial correlations among the features of the drug–microbe pair. For the patches of and , we calculate the spatial attention score matrix , , , both within and between them. Then, we normalize and multiply it with to obtain the spatial correlations representation for drugs from the perspective of self-attention (cross-attention),
| (18) |
Similarly, we compute the spatial correlations representations and for microbes under self-attention and cross-attention. The self-attention focuses on capturing the spatial correlations among the attributes of a drug (microbe) itself. The cross-attention concentrates on reveal the spatial correlations among the attributes of drug node and those of the microbe node. We combine the spatial correlations representations from these multiple perspectives to form and as follows,
| (19) |
where represents element-wise addition. The final output is obtained by concatenating the results of all attention heads,
| (20) |
Subsequently, and are flattened as vectors, stacked vertically, and concatenated to represent the long-distance spatial correlations of node pairs ,
| (21) |
2.6 Multiple representations fusion and optimization
Multiple representations, including the multi-node association representation , homogeneous and heterogeneous neighbor representation , and long-distance spatial correlations representation of and are concatenated to obtain the final node pair representation ,
| (22) |
We employ a convolution-pooling layer (as shown in Fig. 1e) to further integrate . After the l-th convolution-pooling layer, the node pair feature is denoted as ,
| (23) |
where and is activation function. denotes the max-pooling operation and represents the convolution operation. and represent the weight matrix and bias vector of the l-th convolution-pooling layer, respectively. We name the feature obtained from the last convolution-pooling layer as , which enables the prediction of scores p associated with and through the fully connected layer,
| (24) |
where and are the weight matrix and the bias vector, respectively. represents the probability that drug node has (does not have) an association with microbe node . The loss of our model is calculated based on the cross-entropy loss function,
| (25) |
where is the number of nodes per batch of training, and y represents the true association of a drug and a microbe.
3 Experimental evaluations and discussions
3.1 Parameter settings
We trained DHDMP using the Nvidia GeForce RTX 4060, utilizing the PyTorch framework and optimizing it with the Adam algorithm. The training parameters consisted of 80 epochs, a batch size of 32, and a learning rate of 0.0005. The numbers of the encoding layers in NHCN and GCNFP are selected from . We conducted the experiments on all the combinations of layer number of NHCN and that of GCNFP (Supplementary Table S1). When NHCN and GCNFP contain two encoding layers, the model achieves the best prediction performance. Additionally, we established 32 hyperedges to capture complex relationships (Supplementary Table S2). Within the SCA mechanism, the patch size and feature dimension () were set to , with eight attention heads utilized. The fusion of multiple representations involved two convolution layers with kernel sizes of and , respectively, while the pooling layers utilized a window size of . We also studied the impact of datasets with different positive to negative ratio on DHDMP (Supplementary Table S3) and time complexity analysis are provided in Supplementary File S1.
3.2 Evaluation metrics
Five-fold cross-validation is used to evaluate the prediction performance of DHDMP and other comparative methods. All known drug–microbe associations are randomly divided into five equal parts, called positive samples. The number of unknown drug–microbe associations is much larger than the positive examples and these unknown associations are considered negative samples. During the training and testing process of each fold, four parts of positive examples and an equal number of randomly selected negative examples are used as the training set. The remaining positive and negative samples make up the test set.
Evaluation of the models is based on the area under the receiver operating characteristic curve (AUC) (Hajian-Tilaki 2013) and the area under the precision–recall curve (AUPR) (Saito and Rehmsmeier 2015). AUC and AUPR are computed for each fold, and the average of these 5-fold results yields the final AUC and AUPR scores. In addition, the top k recall rate of a microbe-related drug is also calculated, given the importance of identifying microbes associated with drugs for experimental purposes.
3.3 Ablation experiments
In our ablation experiments, we systematically remove components of our model—NHCN, GCNFP, and SCA—to assess their individual contributions to prediction performance (Table 1). When NHCN is excluded, we observe a decline in prediction performance by 2.7% and 13.2% in terms of AUC and AUPR compared to our final model. This highlights the crucial role of capturing the intricate relationships among multiple drugs and microbes in enhancing the accuracy of drug–microbe association prediction. Similarly, without the learning of homogeneous and heterogeneous neighbor representations by GCNFP leads to a decrease in AUC and AUPR by 1.5% and 13.0%, respectively, compared to the complete model. This confirms the significance of incorporating information from both homogeneous and heterogeneous neighbors, which contributes significantly to predicting drug–microbe associations. Furthermore, when SCA is not utilized, DHDMP experiences a reduction of 1.4% in AUC and 6.4% in AUPR. This demonstrates the effectiveness of SCA in capturing the long-distance spatial relationships between node pairs. After the category features were removed from the prediction model, its AUC and AUPR decreased by 1.7% and 5.5%, respectively. It indicated that the category features may reveal the heterogeneity of multiple kinds of nodes. After the node feature propagation was eliminated from the model, its AUC and AUPR decreased by 1.4% and 4.2%. It demonstrates that propagating node information from homogeneous graphs to heterogeneous graph facilitates heterogeneous neighbor feature learning. The results of our ablation experiments indicate that NHCN makes the most contribution to predicting drug–microbe associations. This is primarily attributed to the ability of the hypergraph to effectively represent the complex relationships among multiple drugs and microbes.
Table 1.
Results of ablation studies of DHDMP.
| NHCN | GCNFP | SCA | Category feature | NFPS | Average AUC | Average AUPR |
|---|---|---|---|---|---|---|
| ✗ | ✓ | ✓ | ✗ | ✓ | 0.932 | 0.691 |
| ✓ | ✗ | ✓ | ✓ | ✗ | 0.944 | 0.693 |
| ✓ | ✓ | ✗ | ✓ | ✓ | 0.945 | 0.759 |
| ✓ | ✓ | ✓ | ✗ | ✓ | 0.942 | 0.768 |
| ✓ | ✓ | ✓ | ✓ | ✗ | 0.945 | 0.781 |
| ✓ | ✓ | ✓ | ✓ | ✓ | 0.959 | 0.823 |
The bold values are the highest ones.
3.4 Comparison with other methods
We assess our approach against six leading methods in the field: GCNMDA (Long et al. 2020a), EGATMDA (Long et al. 2020b), GSAMDA (Tan et al. 2022), GACNNMDA (Ma et al. 2023), SCSMDA (Tian et al. 2023), and NGMDA (Xuan et al. 2024). Each of these methods is trained using the optimal parameters mentioned in their original papers. To maintain consistency, we train our model and the comparative methods on the same dataset and use the same partitioning for training and testing. Below, you will find brief descriptions of these six comparative methods.
GCNMDA: This method constructs a network comprising drugs and microbes, leveraging their similarities and associations. By employing GCN and conditional random fields, the model predicts drugs relevant to microbes.
EGATMDA: This approach forms a heterogeneous network covering microbes, diseases, and drugs. It utilizes GCN with node-level attention and graph neural networks with graph-level attention to infer associations between microbes and drugs.
GSAMDA: In this model, drug and microbe similarities are computed using Gaussian interaction profiles (GIP) and Hamming interaction profiles (HIP). The features of drug and microbe nodes are learned through graph attention networks (GAT) and sparse autoencoders.
GACNNMDA: Similarity between drugs and microbes is determined based on GIP and HIP, and association scores between them are computed using convolutional neural networks.
SCSMDA: This method establishes an integrated network of drug and microbe similarities, constructing multiple meta-path-induced networks. It employs a framework based on graph convolution and graph contrastive learning to learn features of drug and microbe nodes.
NGMDA: It builds a drug–microbe heterogeneous graph and enhances node features through fully connected autoencoders. Additionally, a graph transformer is designed to infer associations between drugs and microbes.
Based on the results presented in Table 2, DHDMP achieved the highest average AUC at 95.9%, surpassing GCNMDA by 5.6%, EGATMDA by 1.9%, GACNNMDA by 11.6%, GSAMDA by 5.7%, SCSMDA by 4.3%, and NGMDA by 1.5%. Furthermore, DHDMP’s average AUPR stands at 82.3%, significantly outperforming GCNMDA, EGATMDA, GSAMDA, GACNNMDA, SCSMDA, and NGMDA by 50.8%, 51.6%, 57.6%, 62.7%, 48.3%, and 9.5%, respectively. Our method achieved the highest AUC and AUPR, with NGMDA demonstrating the second-best predictive performance. The superior performance of our method and NGMDA’s AUPR can be attributed largely to the incorporation and utilization of node-pair-level attributes. However, NGMDA falls short compared to our method due to its oversight of the intricate relationships between multiple drugs and microbes. GCNMDA, EGATMDA, and SCSMDA exhibit better performance compared to GSAMDA and GACNNMDA, possible reason is GSAMDA and GACNNMDA only learn node representations based on GATs or GCNs without considering the types of nodes and edges. GCNMDA, EGATMDA, and SCSMDA effectively learn the semantics of connections between drug and microbe meta-path nodes, thus yielding good performance.
Table 2.
Average AUCs and AUPRs of our method and the compared methods over all the 1373 drugs.
| Methods | AUC | AUPR |
|---|---|---|
| GCNMDA | 0.903 | 0.315 |
| EGATMDA | 0.940 | 0.307 |
| GSAMDA | 0.902 | 0.247 |
| GACNNMDA | 0.843 | 0.196 |
| SCSMDA | 0.916 | 0.340 |
| NGMDA | 0.944 | 0.728 |
| DHDMP | 0.959 | 0.823 |
The bold values are the highest ones.
To determine whether our method’s AUC and AUPR are better than those of other methods, we conducted statistical tests. Using 5-fold cross-validation, we calculated the average AUCs and AUPRs for 1373 drugs for each method. Then, we performed Paired Wilcoxon tests to compare the AUCs and AUPRs of DHDMP with those of the other methods. Table 3 shows the statistical results, indicating a significant improvement (P-value <0.05).
Table 3.
Results of the paired Wilcoxon test by comparing DHDMP and any of the other methods.
| P-value | GCNMDA | EGATMDA | GACNNMDA | GSAMDA | SCSMDA | NGMDA |
|---|---|---|---|---|---|---|
| AUC | 8.76e-159 | 5.82e-94 | 1.54e-165 | 1.28e-151 | 6.39e-164 | 3.47e-20 |
| AUPR | 5.25e-188 | 1.33e-162 | 1.78e-196 | 3.31e-190 | 1.18e-212 | 8.12e-33 |
For each drug, we calculated the recall rates of candidate microbes at various top k values (Fig. 4). Our method consistently performs better than other methods for different k values. For example, when k = 3, DHDMP achieves a recall rate of 88.1%, surpassing GCNMDA by 43.4%, EGATMDA by 38.9%, GACNNMDA by 60.9%, GSAMDA by 61.4%, SCSMDA by 43.9%, and NGMDA by 11.5%. In the range of , DHDMP maintains the highest recall rate, while NGMDA closely follows with recall rates of 81.3%, 83.4%, and 85.7%, respectively. EGATMDA emerges as the third-best performing method with recall rates of 67.8%, 74.9%, and 80.1%. GCNMDA’s performance lags slightly behind SCSMDA, achieving recall rates of 61.5%, 66.8%, and 70.8%. While GSAMDA’s recall rates are slightly lower at 55.7%, 63.7%, and 68.4%, they still outperform GACNNMDA by 13.2%, 13.8%, and 12.1%, respectively.
Figure 4.
The average recall rates of drugs at multiple top k cutoffs
3.5 Case studies
We conducted case studies on moxifloxacin, ciprofloxacin, and curcumin to demonstrate DHDMP’s ability to predict microbes that are relevant to these drugs. Moxifloxacin is commonly prescribed for upper and lower respiratory tract infections such as acute sinusitis, pneumonia, and skin and soft tissue infections. Ciprofloxacin is used to treat respiratory tract infections and sepsis, and as a preventive measure against infections in immunocompromised patients. Curcumin, a natural phenolic antioxidant, is known for its potential anti-cancer properties and its possible role in preventing Alzheimer’s disease. Our model was trained using all known associations and an equal number of randomly selected unknown microbe–drug associations. For each drug, the candidate microbes were ranked in descending order according to their association scores. The top 20 candidate microbes for each drug are regarded as its potential candidate microbes. We then analyzed the top 20 candidate microbes for moxifloxacin, ciprofloxacin, and curcumin. The detailed results can be found in Tables 4–6, respectively. The MDAD database provides clinically or experimentally supported associations between microbes and drugs, while the aBiofilm database compiles information on anti-biofilm agents relevant to over 140 microbes (Rajput et al. 2018). These two databases, along with relevant bioinformatics literature, were used to validate the predictions of drug–microbe associations. In the case of moxifloxacin, as listed in Table 4, MDAD contains four potential microbes, three of which are documented in aBiofilm, and evidence supporting 12 candidates can be found in the literature. Notably, studies (Dalhoff et al. 2020, Ontong et al. 2021) indicate that certain microbes such as Escherichia coli, Streptococcus pneumoniae, Staphylococcus aureus, and Klebsiella pneumoniae are inhibited by moxifloxacin, while Listeria monocytogenes and Pseudomonas aeruginosa exhibit resistance to it (Ince et al. 2003, Tahoun et al. 2017, Budzinskaya et al. 2019). Regarding ciprofloxacin, Table 5 reveals that nine of the potential microbes have recently been confirmed to have associations in the literature, 10 are recorded in MDAD, and seven can be found in aBiofilm. Similarly, the results presented in Table 6 for curcumin show that evidence from the aBiofilm and MDAD databases supports 18 of the potential microbes, with one confirmed in the literature.
Table 4.
The top 20 candidate microbes related to moxifloxacin.
| Rank | Microbe name | Evidence | Rank | Microbe name | Evidence |
|---|---|---|---|---|---|
| 1 | Candida albicans | aBiofilm, MDAD | 11 | Salmonella enterica | PMID: 22151215 |
| 2 | Haemophilus influenzae | MDAD | 12 | Vibrio campbellii | Unconfirmed |
| 3 | Stenotrophomonas maltophilia | aBiofilm, MDAD | 13 | Plasmodium falciparum | PMID: 17214980 |
| 4 | Mycobacterium avium | aBiofilm, MDAD | 14 | Influenza B virus | Unconfirmed |
| 5 | Staphylococcus aureus | PMID: 12654680 | 15 | Klebsiella pneumoniae | PMID: 33406110 |
| 6 | Streptococcus mutans | PMID: 29160117 | 16 | Dihydropteroate synthase type-1 | Unconfirmed |
| 7 | Pseudomonas aeruginosa | PMID: 31691651 | 17 | Streptococcus pneumoniae | PMID: 31542319 |
| 8 | Escherichia coli | PMID: 31542319 | 18 | Candida tropicalis | Unconfirmed |
| 9 | Listeria monocytogenes | PMID: 28739228 | 19 | Human immunodeficiency virus 1 | PMID: 18441333 |
| 10 | Burkholderia pseudomallei | PMID: 24502667 | 20 | Streptococcus sanguinis | PMID: 10629010 |
Table 5.
The top 20 candidate microbes related to ciprofloxacin.
| Rank | Microbe name | Evidence | Rank | Microbe name | Evidence |
|---|---|---|---|---|---|
| 1 | Bacillus subtilis | MDAD | 11 | Streptococcus mutans | PMID: 30468214 |
| 2 | Staphylococcus aureus | aBiofilm, MDAD | 12 | Candida albicans | PMID: 31471074 |
| 3 | Escherichia coli | aBiofilm, MDAD | 13 | Staphylococcus epidermidis | PMID: 28481197 |
| 4 | Mycobacterium tuberculosis | MDAD | 14 | Listeria monocytogenes | PMID: 28355096 |
| 5 | Proteus vulgaris | aBiofilm, MDAD | 15 | Enterococcus faecalis | PMID: 27790716 |
| 6 | Haemophilus influenzae | MDAD | 16 | Klebsiella pneumoniae | PMID: 27257956 |
| 7 | Providencia stuartii | aBiofilm, MDAD | 17 | Vibrio campbellii | Unconfirmed |
| 8 | Morganella morganii | aBiofilm, MDAD | 18 | Salmonella enterica | PMID: 26933017 |
| 9 | Pseudomonas aeruginosa | aBiofilm, MDAD | 19 | Streptococcus pneumoniae | PMID: 26100702 |
| 10 | Stenotrophomonas maltophilia | aBiofilm, MDAD | 20 | Vibrio harveyi | PMID: 27247095 |
Table 6.
The top 20 candidate microbes related to curcumin.
| Rank | Microbe name | Evidence | Rank | Microbe name | Evidence |
|---|---|---|---|---|---|
| 1 | Aeromonas hydrophila | aBiofilm, MDAD | 11 | Vibrio vulnificus | aBiofilm, MDAD |
| 2 | Citrobacter freundii | aBiofilm, MDAD | 12 | Raoultella ornithinolytica | aBiofilm, MDAD |
| 3 | Burkholderia cenocepacia | aBiofilm, MDAD | 13 | Staphylococcus aureus | PMID: 32708619 |
| 4 | Candida albicans | aBiofilm, MDAD | 14 | Vibrio parahaemolyticus | aBiofilm, MDAD |
| 5 | Burkholderia multivorans | aBiofilm, MDAD | 15 | Pseudomonas japonica | aBiofilm, MDAD |
| 6 | Vibrio harveyi | aBiofilm, MDAD | 16 | Klebsiella variicola | aBiofilm, MDAD |
| 7 | Proteus mirabilis | aBiofilm, MDAD | 17 | Escherichia coli | aBiofilm, MDAD |
| 8 | Serratia marcescens | aBiofilm, MDAD | 18 | Streptococcus mutans | aBiofilm, MDAD |
| 9 | Enterobacter cancerogenus | aBiofilm, MDAD | 19 | Pseudomonas aeruginosa | aBiofilm, MDAD |
| 10 | Enterobacter ludwigi | aBiofilm, MDAD | 20 | Staphylococcus epidermidis | Unconfirmed |
3.6 Prediction of novel microbe–drug associations
We trained our model using all known drug–microbe associations and an equal number of randomly selected unknown associations. Subsequently, we applied our model to predict candidate microbes for 1373 drugs. The top 20 potential candidate microbes for each drug, as predicted by our model, are listed in Supplementary File S2.
4 Conclusion
We proposed a novel method to integrate the attributes of neighbor nodes in the homogeneous and heterogeneous graphs, and encode the long-distance dependencies within the pairwise attributes for predicting the drug-related microbes. The established hyperedges are beneficial for integrating the complex relationships among multiple drugs and microbes from various perspectives. The hypergraph topology was able to be formed dynamically according to the learnable hyperedge embeddings. The designed NHCNs deeply integrated the complex relationships and the heterogeneity of multi-category nodes. A graph convolutional network with node feature propagation enhanced the feature learning for the drug and microbe nodes by fusing the features from homogeneous and heterogeneous neighbors and their topologies. The presented SCA captured the long-distance spatial correlations among multiple attribute patches of the drug–microbe node pair. The cross-validation experimental results showed DHDMP’s higher AUC and AUPR than the compared six methods. The recall rates for the top-ranked candidates indicated that DHDMP was able to retrieve more actual drug–microbe associations. The case studies on three drugs showed DHDMP’s ability in screening the potential candidate microbes associated with an interested drug.
Supplementary Material
Contributor Information
Ping Xuan, School of Information Science and Engineering, Yanshan University, Qinhuangdao 066004, China; Department of Computer Science and Technology, Shantou University, Shantou 515063, China.
Zelong Xu, School of Information Science and Engineering, Yanshan University, Qinhuangdao 066004, China.
Hui Cui, Department of Computer Science and Information Technology, La Trobe University, Melbourne, VIC 3083, Australia; Australian Centre for AI in Medical Innovation, La Trobe University, Melbourne 3083, Australia.
Jing Gu, School of Computer Science and Technology, Heilongjiang University, Harbin 150080, China.
Cheng Liu, Department of Computer Science and Technology, Shantou University, Shantou 515063, China.
Tiangang Zhang, School of Mathematical Science, Heilongjiang University, Harbin 150080, China.
Peiliang Wu, School of Information Science and Engineering, Yanshan University, Qinhuangdao 066004, China.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
The authors declare that there is no conflict of insterest.
Funding
This work was supported by the Natural Science Foundation of China [62172143, 62372282, 62276028, U20A20167]; Natural Science Foundation of Guangdong Province [2024A1515010176]; Natural Science Foundation of Hebei Province [F202103079]; Natural Science Foundation of Heilongjiang Province [LH2023F044]; Innovation Capability Improvement Plan Project of Hebei Province [22567626H]; and STU Scientific Research Initiation Grant [NTF22032].
References
- Aggarwal N, Kitano S, Puah GRY. et al. Microbiome and human health: current understanding, engineering, and enabling technologies. Chem Rev 2022;123:31–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzinskaya M, Strakhovskaya M, Andreeva I. et al. Conjunctival microflora and its antibiotic sensitivity after serial intravitreal injections. Vestn Oftalmol 2019;135:135–40. [DOI] [PubMed] [Google Scholar]
- Dalhoff A, Bowker K, MacGowan A.. Comparative evaluation of eight in vitro pharmacodynamic models of infection: activity of moxifloxacin against Escherichia coli and Streptococcus pneumoniae as an exemplary example. Int J Antimicrob Agents 2020;55:105809. [DOI] [PubMed] [Google Scholar]
- De Lorenzi-Tognon M, Genton PL, Schrenzel PJ. et al. Synthèse du 8e symposium «feeding the microbiota»: prébiotiques et probiotiques. Rev Med Suisse 2023;19:1149–53. [DOI] [PubMed] [Google Scholar]
- Deng L, Huang Y, Liu X. et al. Graph2MDA: a multi-modal variational graph embedding model for predicting microbe–drug associations. Bioinformatics 2022;38:1118–25. [DOI] [PubMed] [Google Scholar]
- Dosovitskiy A, Beyer L, Kolesnikov A. et al. An image is worth 16x16 words: transformers for image recognition at scale. In: International Conference on Learning Representations, 2021. OpenReview.net.
- Fan L, Wang L, Zhu X.. A novel microbe-drug association prediction model based on stacked autoencoder with multi-head attention mechanism. Sci Rep 2023;13:7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, DeBoy RT. et al. Metagenomic analysis of the human distal gut microbiome. Science 2006;312:1355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med 2013;4:627–35. [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Tanaka N, Kanehisa M. et al. SIMCOMP/SUBCOMP: chemical structure search servers for network analyses. Nucleic Acids Res 2010;38(Suppl 2):W652–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince D, Zhang X, Hooper DC.. Activity of and resistance to moxifloxacin in Staphylococcus aureus. Antimicrob Agents Chemother 2003;47:1410–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipf TN, Welling M. Semi-supervised classification with graph convolutional networks. In: International Conference on Learning Representations, 2017. OpenReview.net.
- Kipf TN, Welling M. Variational graph auto-encoders. stat, 1050:21, 2016.
- Kumbhare SV, Pedroso I, Ugalde JA. et al. Drug and gut microbe relationships: moving beyond antibiotics. Drug Discov Today 2023;28:103797. [DOI] [PubMed] [Google Scholar]
- Li T, Wang Z, Guo J. et al. Bacterial resistance to antibacterial agents: mechanisms, control strategies, and implications for global health. Sci Total Environ 2023;860:160461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Han P, You Z-H. et al. In silico prediction of drug-target interaction networks based on drug chemical structure and protein sequences. Sci Rep 2017;7:11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Li J, Nie R. et al. A graph auto-encoder model for miRNA-disease associations prediction. Brief Bioinform 2021;22:bbaa240. [DOI] [PubMed] [Google Scholar]
- Long Y, Wu M, Kwoh CK. et al. Predicting human microbe–drug associations via graph convolutional network with conditional random field. Bioinformatics 2020a;36:4918–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Wu M, Liu Y. et al. Ensembling graph attention networks for human microbe–drug association prediction. Bioinformatics 2020b;36(Supplement_2):i779–i786. [DOI] [PubMed] [Google Scholar]
- Ma Q, Tan Y, Wang L.. GACNNMDA: a computational model for predicting potential human microbe-drug associations based on graph attention network and CNN-based classifier. BMC Bioinformatics 2023;24:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontong JC, Ozioma NF, Voravuthikunchai SP. et al. Synergistic antibacterial effects of colistin in combination with aminoglycoside, carbapenems, cephalosporins, fluoroquinolones, tetracyclines, fosfomycin, and piperacillin on multidrug resistant Klebsiella pneumoniae isolates. PLoS One 2021;16:e0244673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajput A, Thakur A, Sharma S. et al. aBiofilm: a resource of anti-biofilm agents and their potential implications in targeting antibiotic drug resistance. Nucleic Acids Res 2018;46:D894–D900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Rehmsmeier M.. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 2015;10:e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Bäckhed F.. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 2013;11:227–38. [DOI] [PubMed] [Google Scholar]
- Song W, Xu L, Han C. et al. Drug–target interaction predictions with multi-view similarity network fusion strategy and deep interactive attention mechanism. Bioinformatics 2024;40:btae346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y-Z, Zhang D-H, Cai S-B. et al. MDAD: a special resource for microbe-drug associations. Front Cell Infect Microbiol 2018;8:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahoun AB, Abou Elez RM, Abdelfatah EN. et al. Listeria monocytogenes in raw milk, milking equipment and dairy workers: molecular characterization and antimicrobial resistance patterns. J Glob Antimicrob Resist 2017;10:264–70. [DOI] [PubMed] [Google Scholar]
- Tan Y, Zou J, Kuang L. et al. GSAMDA: a computational model for predicting potential microbe–drug associations based on graph attention network and sparse autoencoder. BMC Bioinformatics 2022;23:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, Yu Y, Fang H. et al. Predicting microbe–drug associations with structure-enhanced contrastive learning and self-paced negative sampling strategy. Brief Bioinform 2023;24:bbac634. [DOI] [PubMed] [Google Scholar]
- Uddin TM, Chakraborty AJ, Khusro A. et al. Antibiotic resistance in microbes: history, mechanisms, therapeutic strategies and future prospects. J Infect Public Health 2021;14:1750–66. [DOI] [PubMed] [Google Scholar]
- VanEvery H, Franzosa EA, Nguyen LH. et al. Microbiome epidemiology and association studies in human health. Nat Rev Genet 2023;24:109–24. [DOI] [PubMed] [Google Scholar]
- Ventura M, O'Flaherty S, Claesson MJ.. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol 2009;7:61–71. [DOI] [PubMed] [Google Scholar]
- Vich Vila A, Collij V, Sanna S. et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun 2020;11:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber KS, Knebel B, Strassburger K. et al. Associations between explorative dietary patterns and serum lipid levels and their interactions with ApoA5 and ApoE haplotype in patients with recently diagnosed type 2 diabetes. Cardiovasc Diabetol 2016;15:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY. et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 2008;455:1109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Guo AC. et al. Drugbank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res 2018;46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Xiong Y, Zhu P. et al. The role of gut microbiota in hypertension pathogenesis and the efficacy of antihypertensive drugs. Curr Hypertens Rep 2021;23:1–14. [DOI] [PubMed] [Google Scholar]
- Xuan P, Gu J, Cui H. et al. Multi-scale topology and position feature learning and relationship-aware graph reasoning for prediction of drug-related microbes. Bioinformatics 2024;40:btae025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R. et al. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019;570:462–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.