Fig. 3.
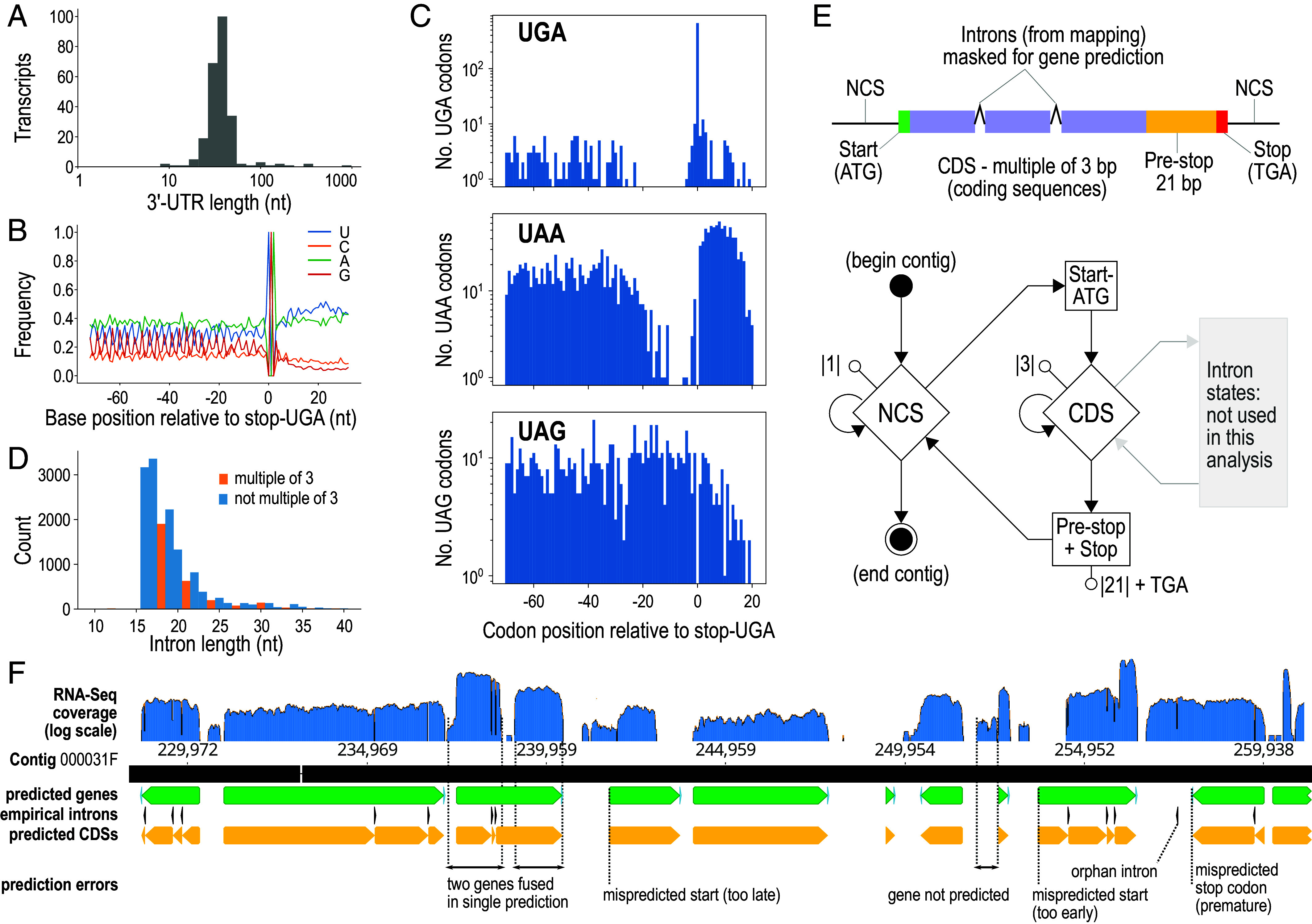
L. magnus gene prediction. (A) Length distribution of 3′-untranslated regions (3′-UTRs) from poly-A-tailed transcripts with stop codons predicted from BLASTX hits to other ciliates. (B) Base composition around predicted stop codons in transcripts. (C) Counts of UGA, UAA, and UAG codons relative to predicted stop-UGA codons in transcripts, showing depletion of in-frame UGA and UAA immediately upstream of stop-UGAs, but no depletion of UAG (cf. SI Appendix, Fig. S7). (D) Length distribution of introns predicted from RNA-seq mapping to MAC assembly (excluding orphan introns). (E) Diagrams of gene model and GHMM used for gene prediction. Start, CDS, and stop states in the GHMM are also mirrored by their corresponding reverse complements. Introns were annotated empirically from RNA-seq mapping. (F) Excerpt of Pogigwasc gene prediction from MAC contig 000031F; annotation tracks for predicted genes (green), CDSs (yellow), empirical introns (black), aligned against RNA-seq coverage (blue). Common types of mispredictions recognizable by comparison with RNA-seq mappings are indicated.
