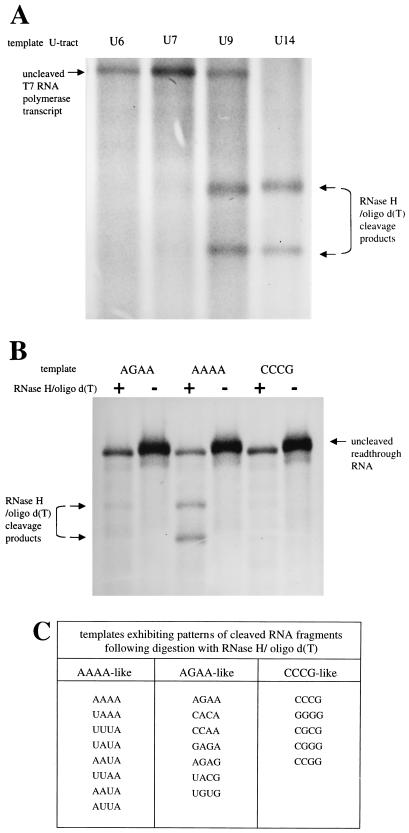
FIG. 2.
Analysis of VSV polymerase slippage by an RNase H-oligo(dT) digestion assay. (A) To determine what length of A tract was cleaved by RNase H in the presence of oligo(dT), positive-strand RNAs were transcribed in BHK cells by T7 RNA polymerase from four plasmids encoding subgenomic templates with gene junction U tracts of 6, 7, 9, and 14 nucleotides. These templates were transcribed to yield positive-stranded RNAs having corresponding A tracts of 6, 7, 9, and 14 nucleotides. Metabolically labeled RNAs were annealed to oligo(dT), digested with RNase H, and then visualized by agarose-urea gel electrophoresis and fluorography. Digestion of the labeled T7 RNA polymerase transcripts with an A6 or A7 tract was insignificant, whereas an expanded tract of nine A's was subjected to extensive digestion, as shown by the presence of two faster-migrating cleavage products. RNAs having A14 tracts were digested to completion. (B) Metabolically labeled RNAs transcribed by the VSV polymerase from three subgenomic templates were subjected to the RNase H-oligo(dT) cleavage assay and visualized by agarose-urea gel electrophoresis and fluorography. The tetranucleotide sequences of gene junctions of these templates are shown above the corresponding lanes, written 3′-5′; + and − denote presence and absence of oligo(dT). As described above, cleavage products following RNase H/oligo(dT) digestion were released only when the A tract contained more than seven residues. This assay demonstrated that VSV polymerase slippage was more prevalent on RNAs transcribed from the subgenomic template with the tetranucleotide sequence 3′-AAAA-5′ than on templates with tetranucleotide sequence 3′-AGAA-5′ or 3′-CCCG-5′. (C). RNAs transcribed from a total of 20 templates with altered tetranucleotide sequences were analyzed by the RNase H/oligo(dT) slippage assay, and the results were tabulated according to a qualitative assessment of whether digestion of the corresponding RNAs resembled the digestion pattern of RNAs transcribed from the three templates shown in panel 2B. AAAA-like corresponds to abundant digestion, CCCG-like corresponds to no detectable digestion, and AGAA-like corresponds to an intermediate level of digestion.