TABLE 2.
Sequence of recombinant junctions in molecules recovered from cells transfected with linear substrates encoding sequence-tagged (mismatched) terminia
| Sequence-tagged mismatches at homologous endsb | Fraction of recovered molecules with sequence Xc for nucleotide:
|
Predicted effect of resection polarity on junction sequence
|
||||
|---|---|---|---|---|---|---|
| G | A | T | C | 3′ to 5′ | 5′ to 3′ | |
| Left end | ||||||
 |
5/16 | 3/16 | 6/16 | 2/16 | G or T | A or C |
| Right end | ||||||
 |
5/16 | 8/16 | 1/16 | 2/16 | G or A | T or C |
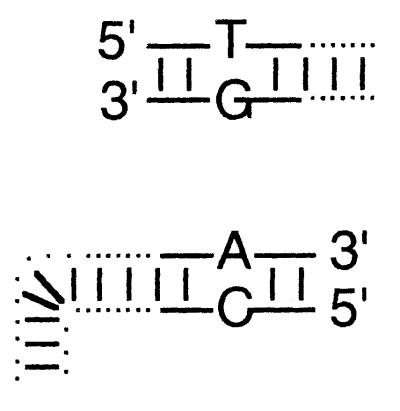
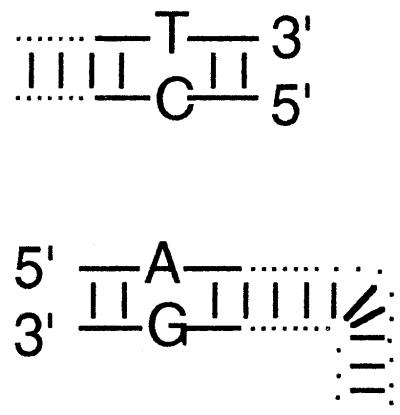
Duplex oligonucleotides were ligated to all four ends of two linear recombination substrates, adding 20 nt of sequence identity to each end interrupted by a single, central, mismatched base pair. These substrates were transfected into virus-infected cells (Fig. 3), and recombinants were recovered in E. coli (6) and sequenced (25).
Diagrams illustrate the base substitutions used to differentiate all eight 3′- and 5′-ended strands. The left end of the linear insert is identical to the vector end shown immediately below it, except that T · G and A · C mismatches differentiate insert from vector ends. T · C and A · G mismatches similarly differentiate the right end of the insert from its vector target. See Materials and Methods for sequence details.
Top-strand sequence (as drawn) at variant site.
