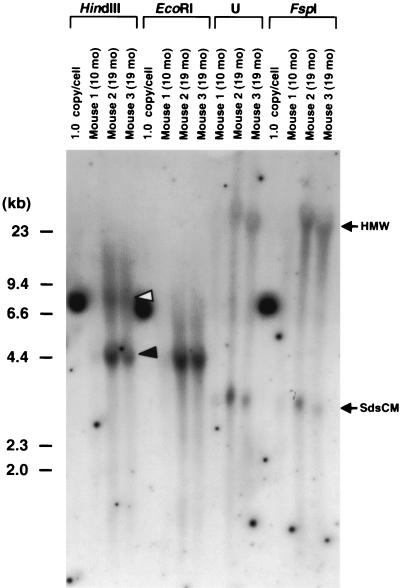
FIG. 2.
Southern blot analysis of liver DNA from three mice injected via the portal vein with AAV-EF1α-F.IX at a dose of 2.7 × 1011 vg per animal. Twenty micrograms of total mouse liver DNA was digested with the enzymes indicated above the lanes or undigested (U), separated on an 0.8% agarose gel, blotted on a nylon membrane, and hybridized with a vector-specific PpuMI EF1α-hF.IX probe (the same as probe C in reference 27). The time of sacrifice is shown above each lane. The 1.0-copy/cell standard is 20 μg of naive mouse liver DNA mixed with the appropriate number of plasmid pV4.1e-hF.IX molecules. This control plasmid is linearized when cut with HindIII, EcoRI, or FspI, generating a 7.6-kb band. HindIII and EcoRI cut the vector genome once at the 3′ side and the center of the vector genome, respectively, while FspI does not cut within the vector genome. Head-to-tail and head-to-head molecules are denoted by closed and open arrowheads, respectively. The positions of high-molecular-weight (HMW) species and supercoiled ds circular monomers (SdsCM) of vector genomes are shown. Ethidium bromide staining of the gel showed no lane-to-lane variations of the amount of loaded DNA among digestions. Note that extrachromosomal forms of the vector genomes (SdsCM) are readily detectable. The Southern blot results for mice sacrificed 3 months postinjection were previously published (27).