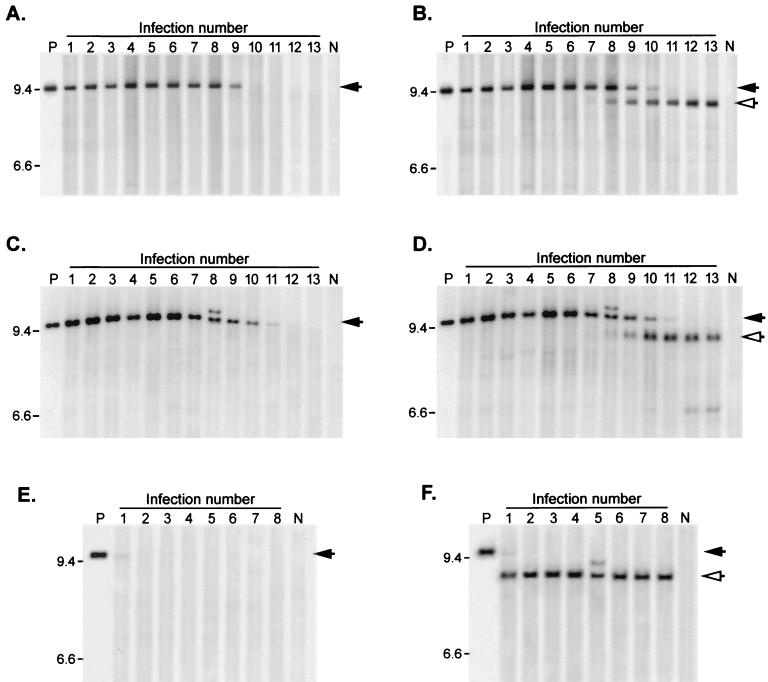
FIG. 3.
Stability of insert-containing genomes over multiple serial infections. Viruses were subjected to repeated serial passage through NIH 3T3 cultures as described in Materials and Methods. Unintegrated proviral DNA was isolated from each virus passage, digested with NheI, and subjected to Southern blotting. (A and B) DNA from ZAPd-puro serial infections, using pac-specific probe (A) or MLV LTR-gag probe (B). (C and D) DNA from ZAPd-GFP serial infections, using GFP-specific probe (C) or LTR-gag probe (D). (E and F) DNA from ZAPd-hygro serial infections, using hph-specific probe (E) or LTR-gag probe (F). Lanes P, NheI-digested plasmid DNA encoding the corresponding virus. Lanes N, DNA isolated from mock-infected cells. Intact full-length provirus signals are indicated by solid arrows, and deletion mutants are indicated by open arrows. Expected full-length fragment sizes: ZAPd-puro, 9,437 bp; ZAPd-GFP, 9559 bp; ZAPd-hygro, 9851 bp. Positions of size standards are shown on the left (in kilobases).