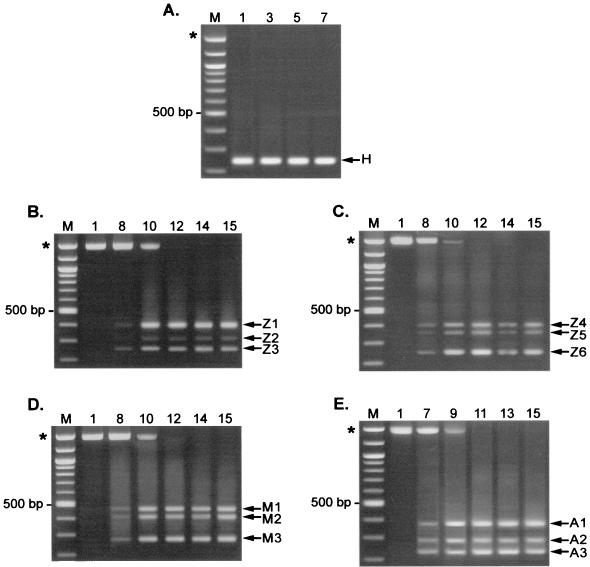
FIG. 5.
PCR analysis of transgene insert region of serially passaged viruses. Hirt DNA from each indicated infection cycle was used as the template in PCR using an upstream primer specific for the appropriate env gene and a common downstream primer specific for the 3′ UTR-3′ LTR border region. (A) Amplification of passaged ZAPd-hygro. Overexposure of this gel (not shown) revealed the presence of a faint band of approximately the size expected for full-length ZAPd-hygro only in cycle 1. (B and C) Amplification of proviral DNA from two independent infection series using ZAPd-GFP. (D) Passaged ZAPm-GFP. (E) Passaged AZE-GFP. Lanes M, 100-bp molecular size markers. Asterisks indicate size of product expected for undeleted, full-length virus.