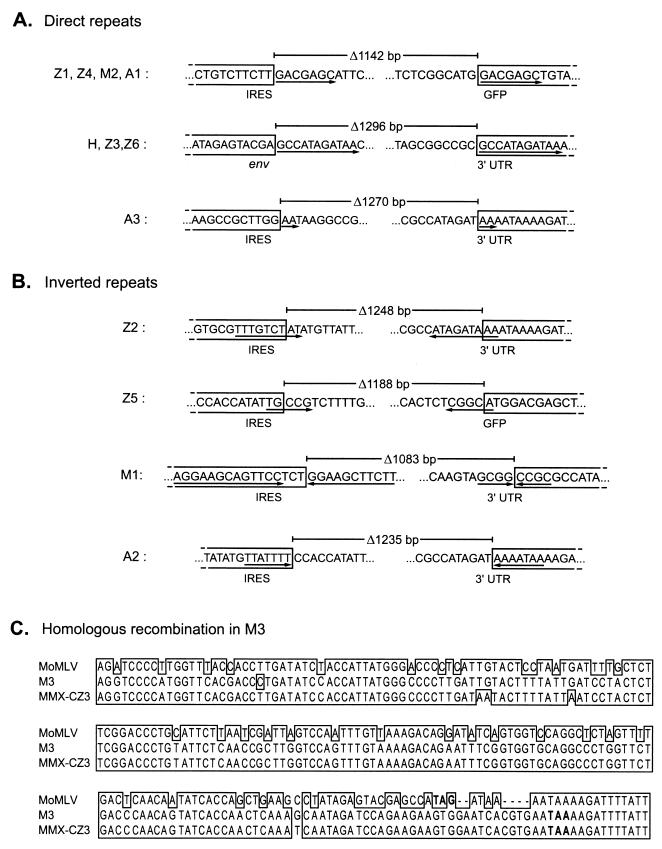
FIG. 6.
Sequence analysis of deletion junctions of variants identified by PCR. Each of the PCR products shown in Fig. 5 was purified and sequenced. The sequences remaining in each deletion variant are boxed, and the intervening sequences deleted from the parental virus are unboxed. Lengths of deletions are indicated above each variant. (A) Deletion junctions of variants whose deletions were flanked by direct repeats in the parental genome. Direct repeats are underlined with arrows. (B) Deletion junctions characterized by inverted repeats in parental virus. Inverted repeats are underlined with arrows, and the orientation of each repeat is indicated by arrow direction. (C) Alignment of ZAPm-GFP variant M1 with Moloney MLV and endogenous virus MMX-CZ3 by the Clustal X program (23). Envelope stop codons are indicated in bold.