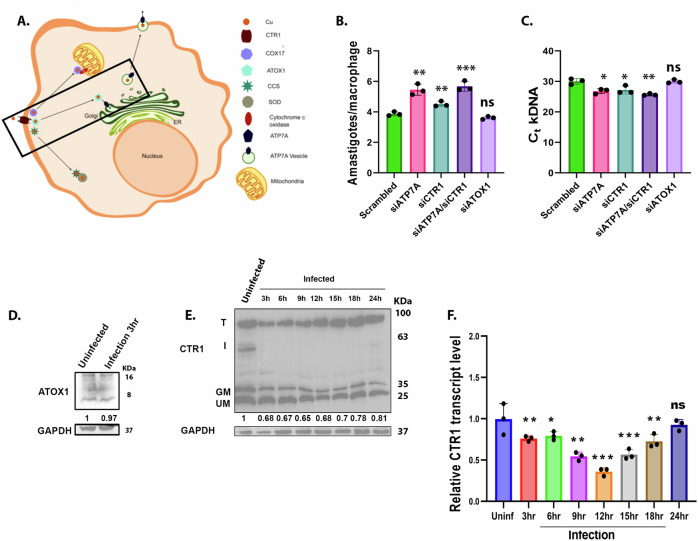
Fig. 4. Knocking down of host ATP7A and CTR1 enhances the infectivity of the Leishmania pathogen.
A Illustration depicting the mammalian copper uptake and utilization pathway. Copper secretory pathway is highlighted. Illustration is prepared in Inkscape 1.2.2. B J774A.1 macrophages, after treatment with scrambled, ATP7A, CTR1 and ATOX1 siRNA, were infected with L. major. After 12 h, amastigote counts inside the macrophage are plotted. At least 100 macrophages were counted from triplicate experiments. Error bars represent mean ± SD of values from three independent experiments. Asterisks indicate values that are significantly different from Scrambled samples. ∗p ≤ 0.05, ∗∗∗∗p ≤ 0.0001; (Student’s t test). C J774A.1 macrophages, after treatment with scrambled, ATP7A, CTR1 and ATOX1 siRNA, were infected with L. major. After 12 h, Ct values of L. major kDNA are plotted. Error bars represent mean ± SD of values from three independent experiments. Asterisks indicate values that are significantly different from Scrambled samples. ∗∗∗∗p ≤ 0.0001, ns; (Student’s t test). D Immunoblot of ATOX1 at 3 h time point after infecting J774A.1 macrophages with L. major promastigotes. The fold change of ATOX1 abundance normalized against housekeeping control GAPDH has been mentioned. E Immunoblot of CTR1 at indicated time points after infecting J774A.1 macrophages with L. major promastigotes. T denotes CTR1 trimer, I denotes Intermediate band, GM denotes glycosylated monomer, UM denotes unglycosylated monomer. The fold changes of CTR1 glycosylated monomer abundance normalized against housekeeping control GAPDH have been mentioned. GAPDH loading control is same for Figs. 2A and 4E where same conditions were used to check the protein levels of ATP7A and CTR1. F qRT–PCR shows CTR1 transcript level at indicated time points after infection, normalized against uninfected control and housekeeping control GAPDH mRNA levels. Error bars represent mean ± SD of values calculated from three independent experiments. Asterisks indicate values that are significantly different from Uninf samples.∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ns; (Student’s t test).