Fig. 2. Efficiency of the methylation-sensitive restriction enzymes.
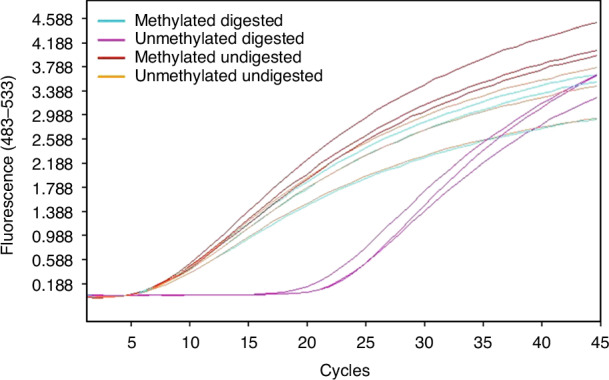
Amplification curve of qPCR of (un)methylated and (un)digested lambda DNA. This is an example of one primer pair hybridizing around a MSRE recognition site. Two types of control samples were used, unmethylated lambda DNA and artificially methylated lambda DNA by CpG methyltransferase M.SssI (New England Biolabs). Each sample was amplified in triplicate. Methylated digested (blue), unmethylated digested (purple), methylated undigested (red) and unmethylated undigested (orange) samples have an average Ct value of 5.7, 22.0, 5.0 and 5.0, respectively.
