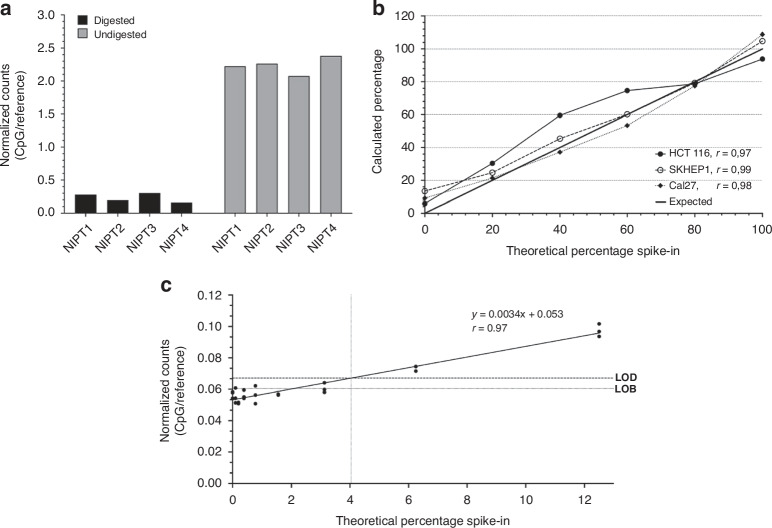
Fig. 7. Potential for liquid biopsies.
a Normalized counts of digested and undigested cfDNA samples. Four cfDNA samples were used to verify that the protocol works in liquid biopsies. On the left side, digested samples are shown. On the right side, undigested samples are shown as positive controls. The DNA input amount is 5 ng. b Calculated percentages based on normalized counts for all target sites of normal cfDNA with spiked-in cell line DNA. Sheared cell line DNA was spiked into normal cfDNA at 0-20-40-60-80-100%. The percentages are calculated by the linear regression through the normalized counts for each sample. The expected value ranges from 0% to 100%, although it is known that normal samples are not 0% methylated in all our targets. The correlation of the calculated and expected percentages is given by r for each cell line. See Supplementary methods for calculations. c Limit of methylation detection calculated by the normalized counts for all target sites. Fully methylated human DNA was spiked into unmethylated human DNA at 0-0.10-0.20-0.39-0.78-1.56-3.13-6.25-12.5% in triplicate. The linear regression through the normalized counts for each sample has a Pearson correlation coefficient of r = 0.97. The limit of blank (LOB) is 0.060 and the limit of detection (LOD) is 0.067. The methylation percentage corresponding to the LOD is 4.04%. Linear regression and plotting was performed using GraphPad Prism.