Abstract
Purpose:
Avelumab (anti–PD-L1) became the first approved treatment for metastatic Merkel cell carcinoma (mMCC) based on results from the phase II JAVELIN Merkel 200 trial. In this study, we report exploratory biomarker analyses from the trial.
Patients and Methods:
Patients with mMCC (n = 88) with or without prior first-line chemotherapy received avelumab 10 mg/kg every 2 weeks. We conducted analyses on somatic mutations, mutational signatures, and tumor mutational burden using paired whole-exome sequencing. Additionally, we examined gene and gene set expression, immune content from RNA sequencing profiles, as well as tumor PD-L1 and CD8 statuses from IHC and CD8 status from digital pathology.
Results:
Tumors positive for Merkel cell polyomavirus (MCPyV) were characterized by an absence of driver mutations and a low tumor mutational burden, consistent with previous studies. A novel MCPyV-specific host gene expression signature was identified. MCPyV+ tumors had increased levels of immunosuppressive M2 macrophages in the tumor microenvironment, which seemed to correlate with PD-L1 expression; high CD8+ T-cell density in these tumors did not predict response to avelumab. Conversely, in patients with MCPyV− tumors, higher CD8+ T-cell density seemed to be associated with response to avelumab. Mutations in several genes were associated with treatment outcomes. Compared with tumors sampled before chemotherapy, tumors sampled after chemotherapy had downregulated gene signatures for immune responses, including reduced expression of IFNγ-related pathways. Levels of activated dendritic cells in responding patients were higher in patients assessed after versus before chemotherapy.
Conclusions:
Exploratory analyses provide insights into mMCC biology and potential associations with response to avelumab. Chemotherapy seems to negatively modulate the immune microenvironment.
Translational Relevance.
Associations between biomarkers and response to immune checkpoint inhibitors in metastatic Merkel cell carcinoma (mMCC) are not fully understood. We report exploratory biomarker analyses from cohorts of patients with treatment-naïve or chemotherapy-treated mMCC who received avelumab (anti–PD-L1) in the JAVELIN Merkel 200 trial. To our knowledge, this is the largest mMCC cohort to be characterized with whole-exome sequencing, paired RNA sequencing profiles, and other technologies. Our findings extend previous studies by showing different biomarker profiles and an immunosuppressive microenvironment in tumors positive for Merkel cell polyomavirus, which are known to have a low tumor mutational burden. In contrast, tumorigenesis in Merkel cell polyomavirus-negative tumors is driven by genomic changes. Samples taken before versus after chemotherapy in different patients showed immunosuppressive effects associated with prior chemotherapy, suggesting that immunotherapy with avelumab may have greater potential for clinical benefits when administered as first-line therapy for mMCC.
Introduction
Merkel cell carcinoma (MCC) is a rare and aggressive skin cancer that is associated with the presence of clonally integrated Merkel cell polyomavirus (MCPyV) in most cases, with additional risk factors including exposure to UV radiation and immunosuppression (1). MCPyV encodes two viral oncogenes (the small and large T antigens) that are critical for tumorigenesis (2). MCC tumors associated with UV exposure (i.e., MCPyV− tumors) are characterized by high tumor mutational burden (TMB) and mutations in TP53 and RB1 (3, 4). Previous studies have shown distinct RNA expression profiles and prognostic biomarkers for MCPyV+ versus MCPyV− tumors (5, 6).
In 2017, avelumab (anti–PD-L1) became the first approved treatment for patients with metastatic MCC (mMCC). Avelumab was approved in any treatment line based on the occurrence of durable responses in patients enrolled in the JAVELIN Merkel 200 trial (7). In patients with mMCC who received avelumab as second-line or later treatment after disease progression with prior chemotherapy [part A (N = 88); referred to as the 2L+ cohort], the objective response rate (ORR) was 33.0% [95% confidence interval (CI), 23.3%–43.8%; ref. 8]. In patients who received first-line avelumab treatment [part B (N = 116); referred to as the 1L cohort], the ORR was 39.7% (95% CI, 30.7%–49.2%; ref. 9). In both cohorts, responses were seen regardless of tumor PD-L1 or MCPyV status. However, objective responses were observed in numerically higher proportions of patients with MCPyV− versus MCPyV+ tumors, those with PD-L1+ versus PD-L1− tumors, and those who received avelumab as 1L versus 2L+ treatment (of note, the proportion of patients with PD-L1+ tumors was lower in the 1L versus the 2L+ cohort).
Initial exploratory biomarker analyses have been reported from the trial (8, 9). In both 1L and 2L+ cohorts, TMB was higher in MCPyV− versus MCPyV+ tumors. In addition, tumors of responders tended to have enrichment of gene expression for pathways associated with inflammatory responses (e.g., IFNγ) and higher CD8+ T-cell density at the invasive margin, with the latter associated with higher MHC class I gene expression. In the 1L cohort, differential gene expression (number of genes differentially expressed) was greatest in tumors with differing MCPyV status (presence or absence) or CD8+ T-cell density. In the 2L+ cohort, responders and patients with MCPyV− tumors had the highest gene set enrichment analysis scores for IFNγ response, TNFα signaling via NF-κB, NK cell activation, and p53 pathway gene sets (8, 9). In biomarker analyses from studies of immune checkpoint inhibitors other than avelumab in MCC, changes observed during treatment included increased densities of intratumoral CD4+/PD-1+ and CD8+/PD-1+ cells and increased proliferation of CD8+ cells. Observations in responders included increased proportions of CD79+ B-lineage cells, decreased densities of Ki67+ (proliferating) tumor cells, and higher densities of PD-1+ and PD-L1+ cells; no association between CD8+ density or infiltration and response was observed (10–12).
The relationship between biomarkers and response to immune checkpoint inhibitors requires further exploration in various tumor types, including mMCC (13). Although durable responses have been reported in a subset of patients with mMCC treated with avelumab (8, 9, 14) or other immune checkpoint inhibitors (15), no single biomarker has been consistently associated with clinical benefit. In mMCC, analyses of gene expression and immune cell profiles are hampered by small data sets and low ORR in some cohorts. Previous MCC genomic studies based on panel-based or targeted gene sequencing to measure genomic changes have been reported (16–18); however, limited data are available from full transcriptomic profiles.
In this study, using the largest mMCC population to be characterized with whole-exome sequencing (WES) and paired RNA sequencing (RNA-seq) profiles to date, in addition to digital pathology analysis, we performed a range of exploratory analyses to investigate the potential relationships between MCPyV status, mutation rate, CD8+ T-cell density, inflammatory pathways, treatment line, and response in patients from both cohorts of the JAVELIN Merkel 200 trial. We also analyzed biomarker expression in publicly available single-cell RNA-seq data from patients with MCPyV+ mMCC (19). To our knowledge, we report the first integrative biomarker analysis to compare cohorts of patients with treatment-naïve versus previously treated mMCC, thereby evaluating the potential impact of chemotherapy on biomarker profiles and response to immune checkpoint inhibitor treatment.
Patients and Methods
Study design
The study design of the phase II, single-arm, open-label, multicenter JAVELIN Merkel 200 trial (NCT02155647) has been reported previously (9, 14). Briefly, eligible patients had histologically confirmed, measurable (per RECIST version 1.1), stage IV MCC; were ≥18 years of age; had an Eastern Cooperative Oncology Group performance status of 0 or 1; and had adequate hematologic, hepatic, and renal functions. In part A, patients had disease progression following ≥1 prior line of chemotherapy for metastatic disease (2L+ cohort). In part B, patients had received no prior systemic therapy for metastatic disease (1L cohort). Previous therapy with any immune checkpoint inhibitor was not permitted in either cohort.
In the 2L+ cohort, the primary endpoint was confirmed best overall response per RECIST 1.1 by an independent review committee. In the 1L cohort, the primary endpoint was durable response, defined as objective response (complete or partial response) with a duration ≥6 months per RECIST 1.1 by an independent review committee. Other endpoints included duration of response, overall survival (OS), progression-free survival (PFS), and safety and tolerability.
The trial was conducted in accordance with the Declaration of Helsinki and the International Council for Harmonisation Guidelines on Good Clinical Practice. The protocol was approved by the independent ethics committee or institutional review board at each participating center, and all patients provided written informed consent before enrollment.
Treatments and assessments
Patients received avelumab 10 mg/kg intravenously every 2 weeks until confirmed disease progression, unacceptable toxicity, or other criteria for withdrawal occurred. Tumors were assessed radiologically every 6 weeks per RECIST 1.1, with adjudication by an independent review committee. Clinical activity was assessed in all patients who received ≥1 dose of avelumab. ORR were calculated with two-sided 95% CI using the Clopper–Pearson method.
Biomarker analyses
IHC and next-generation sequencing analyses used formalin-fixed, paraffin-embedded tumor samples obtained from the metastatic site, collected before receiving avelumab. Formalin-fixed, paraffin-embedded tumor samples were stored at −20°C. PD-L1 status was assessed using the PD-L1 IHC 73-10 pharmDx assay (Dako), and PD-L1+ status was defined as expression in ≥1% of tumor cells. MCPyV status was assessed by IHC using a mouse IgG2b mAb (clone CM2B4, 5 μg/mL; Santa Cruz Biotechnology; RRID: AB_2013156) with staining and detection performed on a BenchMark XT autostainer. MCPyV IHC staining intensity was assessed by trained pathologists and evaluated using four categories (negative, weak, moderate, and strong), with MCPyV+ samples showing unequivocal strong nuclear staining of MCC cancer cells and MCPyV− status being samples that did not fulfill the criteria for positive staining. MCPyV status was also assessed by RT-PCR using DNA extracted from tumor samples, TaqMan reagents (Thermo Fisher Scientific), and small T antigen–specific primers.
CD8 IHC was performed using a mouse mAb (clone C8/144B; Dako; RRID: AB_3073940) and mouse IgG1ϰ (clone MOPC-21, Abcam; RRID: AB_2736846) negative control reagent antibody utilizing a Dako autostainer (Model Link 48) and following validated protocols (Mosaic Laboratories, MOS354-APD). Glass slides were scanned using the Aperio ScanScope AT Turbo system (Aperio). CD8 IHC expression was evaluated by digital image analysis using the Aperio Nuclear V.9 algorithm (Leica Biosystems) and image analysis algorithms developed in Definiens Developer (AstraZeneca). The digital imaging assays produced tabular data at the cellular, region of interest, and slide levels. CD8+ T-cell density measurements were derived from the count of all CD8+ cells compared with the slide-level area analyzed.
Paired RNA-seq and WES next-generation sequencing profiles from patients enrolled in both cohorts were generated by Personalis (Personalis, Inc.). Paired normal WES profiles from blood were available for most tumor samples sequenced. Sample quality control metrics included total yield (≥100 ng RNA/200 ng DNA) and sample integrity as measured by Agilent RNA TapeStation (RNA; ≥30 cutoff) and fluorimetry (DNA; DNA integrity number ≥3). Library preparation was performed using DNA HyperPrep and Stranded RNA-seq kits (Kapa Biosystems); sequencing was performed using Personalis Accuracy and Content Enhanced (ACE) ImmunoID enrichment of a proprietary panel of targeting probes (20, 21) and the Illumina NovaSeq 6000 instrument. The Personalis ACE Cancer Exome sequencing analysis pipeline, which uses Burrows–Wheeler Aligner (RRID: SCR_010910), GATK (RRID: SCR_001876), MuTect (RRID: SCR_000559), VarDict (RRID: SCR_023658), and Picard (RRID: SCR_006525), was applied to generate variant calls. Variant calls were further filtered to remove germline variants found in normal tissues. Mutations with a minimum of five mutant reads (i.e., found in ≥5 separate DNA molecules in an individual tumor sample) that were not annotated as synonymous variants and annotated as resulting in a change in protein-coding sequences, were included in the analysis. Transcript levels were quantified using the Personalis ACE Cancer Transcriptome Analysis pipeline, which uses STAR (RRID: SCR_004463) version 2.4.2a-p1 to align reads to the NCBI hs37d5 annotation 105 reference genome and produces transcripts per million values for each gene. All statistical analyses and hypothesis testing were carried out using R (R Foundation for Statistical Computing; RRID: SCR_001905) and Bioconductor (RRID: SCR_006442) packages. Differential gene expression analysis was performed using the limma package (RRID: SCR_010943) after voom-transforming the RNA-seq counts. Gene set enrichment analysis was performed using the fgsea package (RRID: SCR_020938) with the Molecular Signatures Database (Broad Institute; RRID: SCR_016863), and other gene sets were used with ranked gene lists. Differential analyses were run with parametric (e.g., t test) or nonparametric (e.g., Wilcoxon) tests as appropriate. Immune content deconvolution, including the analysis of M2 macrophage infiltration, was performed using xCell (https://xcell.ucsf.edu/), CIBERSORT (Stanford University; RRID: SCR_016955), and an internal collection of immune-related signatures. To test the impact of prior therapy (treatment line) on response, an ANOVA F test was devised between two multivariate models that corrected for important baseline clinical covariates.
To assess the relationship between mutations and survival or response, two sets of mutations were analyzed: all somatic mutations (from the Personalis output) and mutations only within protein-coding regions.
Kaplan–Meier plots showing associations between mutations and PFS or OS were generated for genes that were mutated in ≥5 patients and for which the P value was ≤0.01 (not corrected for multiple testing). Associations between mutations and responses were also examined.
All analyses were exploratory, hypothesis-generating, and were based on a small cohort but genome-scale dataset. The study was not designed to assess statistical significance in any of the analyses being reported. Analyses were subjected to both the type I (false-positive) error due to multiple comparisons and type II (false-negative) error due to lack of statistical power for the test of interaction (22). To reduce potential type II errors, the results of statistical analyses are presented without correction for multiple comparisons.
Analysis of single-cell RNA-seq data
Quantitated single-cell expression data from Paulson and colleagues (19) were downloaded from Gene Expression Omnibus (GSE117988). Data analysis was conducted using the Seurat Bioconductor package (RRID: SCR_016341). Marker information from the publication and Azimuth (Human Biomolecular Atlas Program; RRID: SCR_021084) was used to annotate cell types.
Data availability
The data generated in this study are available upon request from the sponsor. Any requests for data by qualified scientific and medical researchers for legitimate research purposes will be subjected to the healthcare business of Merck KGaA, Darmstadt, Germany's (CrossRef Funder ID: 10.13039/100009945) Data Sharing Policy. All requests should be submitted in writing to the data sharing portal of the healthcare business of Merck KGaA, Darmstadt, Germany's data sharing portal (https://www.emdgroup.com/en/research/our-approach-to-research-and-development/healthcare/clinical-trials/commitment-responsible-data-sharing.html). When the healthcare business of Merck KGaA, Darmstadt, Germany has a coresearch, codevelopment, or comarketing or copromotion agreement or when the product has been out-licensed, the responsibility for disclosure might be dependent on the agreement between parties. Under these circumstances, the healthcare business of Merck KGaA, Darmstadt, Germany will endeavor to gain agreement to share data in response to requests.
Results
Patients and samples
WES and paired RNA-seq profile data available from 88 patients enrolled in JAVELIN Merkel 200 (2L+, n = 38; 1L, n = 50) were analyzed. Data cutoff dates were September 24, 2017, for the 2L+ cohort (≥2 years of follow-up) and May 2, 2019, for the 1L cohort (≥15 months of follow-up). Baseline characteristics in evaluable patients with WES and RNA-seq samples were generally consistent with those of the overall trial population (Supplementary Table S1) and were representative of the overall patient population with MCC (Supplementary Table S2). Numerical differences between biomarker-evaluable patient sets in the 2L+ versus 1L cohorts included PD-L1+ status (76.3% vs. 22.0%), MCPyV+ status (73.7% vs. 54.0%), and the presence of visceral metastasis (52.6% vs. 68.0%). In the 2L+ cohort, tumor samples were obtained before chemotherapy (i.e., at diagnosis) in 20 patients and after chemotherapy in 18 patients; no paired samples (i.e., pre- and postchemotherapy) were available. The most commonly received prior (1L) chemotherapy agents were etoposide, cisplatin, and carboplatin. The number of events for prognostic outcomes in the two cohorts is shown in Supplementary Table S3.
Characterization of somatic mutations
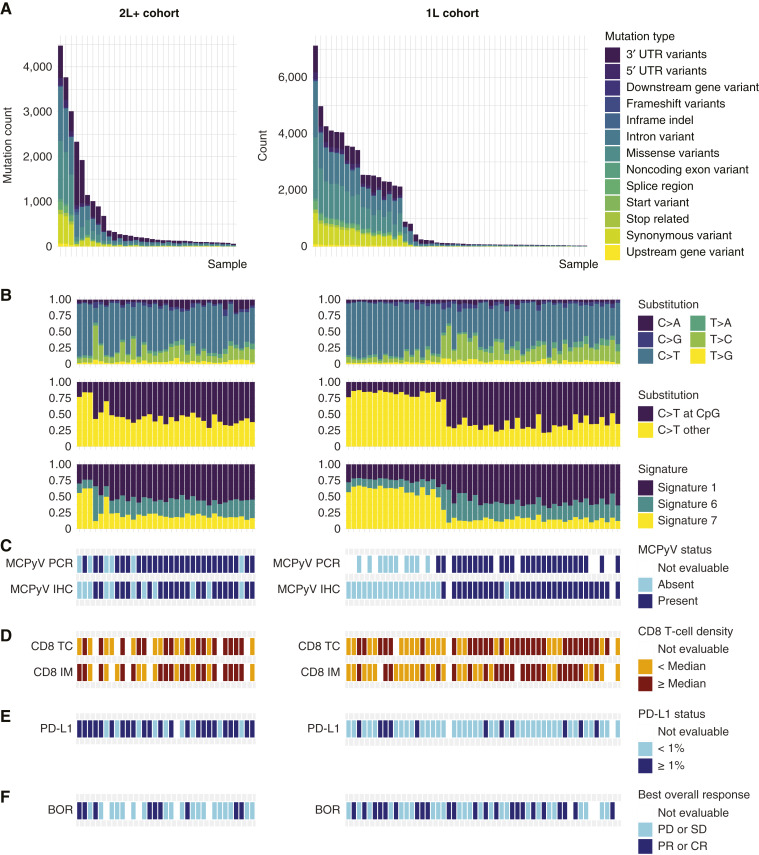
In both cohorts, high mutation rates and driver gene mutations seemed to be associated with the absence of MCPyV (Fig. 1), consistent with previous studies. Clusters of mutation signatures were similar between the cohorts; however, the total numbers were different, with mutation signatures 1, 6, and 7 highly prevalent in both cohorts (Supplementary Fig. S1; ref. 23). Of the top 32 most frequently mutated genes in each cohort, seven genes were commonly mutated in both sets of samples (XIRP2, TTN, TP53, RYR2, RB1, MUC16, and CSMD1). As expected, mutations in cancer drivers were generally absent in MCPyV+ tumors (Supplementary Fig. S2). Mutations in six genes had associations with lack of response with close to nominal significance in patients from the 1L cohort with MCPyV− tumors: RB1, PCLO, NEB, ROS1, RELN, and CPS1 (Supplementary Table S4).
Figure 1.
Characterization of samples from the 2L+ and 1L cohorts (left and right, respectively). A, Occurrence of somatic mutation types. B, Frequency of substitutions and selected gene expression signatures (23). C, MCPyV status. D, CD8+ T-cell density. E, PD-L1 status. F, Best overall response. BOR, best overall response; CR, complete response; IM, invasive margin; PD, progressive disease; PR, partial response; SD, stable disease; TC, tumor core; UTR, untranslated region.
In both cohorts, high CD8+ density was observed in patients whose tumors were MCPyV+ and had a low mutation rate (Fig. 1C and D). Interestingly, a different mutation and gene expression profile was observed in samples collected postchemotherapy from two patients in the 2L+ cohort who had MCPyV+ tumors by PCR and IHC but whose tumors had a high TMB. Using the number of total mutations, TMB, and signatures 1 (age) and 7 (UV exposure), MCPyV status could be reliably predicted using AUC (≥0.7; Table 1). The lower predictive ability in samples from the 2L+ cohort versus those from the 1L cohort was likely due to the two patients with MCPyV+ tumors with distinct mutation profiles.
Table 1.
AUC for predicting MCPyV status. Mutation signatures 1 and 7 were described by Alexandrov and colleagues (23).
| AUC | 2L+ cohort | 1L cohort | ||
|---|---|---|---|---|
| PCR | IHC | PCR | IHC | |
| Total mutations | 0.77 | 0.71 | 1 | 0.98 |
| TMB | 0.77 | 0.67 | 1 | 0.99 |
| Mutation signature 1 | 0.80 | 0.64 | 1 | 0.97 |
| Mutation signature 7 | 0.70 | 0.70 | 1 | 0.98 |
In both cohorts combined, patients with mutations in any region of ATXN3, CACNA1A, IL-17A, MYO7B, and TRRAP had shorter OS and PFS. In addition, patients with mutations in the protein-coding region of CTNND2 had shorter OS and PFS (particularly in patients with MCPyV− tumors), and those with mutations in the protein-coding regions of PKHD1 and GRIN2A had shorter PFS (Supplementary Fig. S3). In analyses of associations between gene mutations and response in the combined 1L and 2L+ cohorts, genes of interest included GRIN2A, IRAK2, KIF5C, LYST, and MYO7B; however, potential associations were seen with mutations in many other genes, particularly within the 1L cohort. In tumors from patients in the 1L MCC cohort, mutations in CTNND2, unlike mutations in other WNT/β-catenin pathway genes, did not impact WNT/β-catenin pathway transcription (Supplementary Fig. S4A and S4B). However, patients with CTNND2 mutations had lower IFNγ expression (Supplementary Fig. S4C), and all patients had progressive disease as the best response. Gene pathways differentially enriched between subgroups defined by MCPyV or response status are shown in Supplementary Fig. S5.
Expression profile by MCPyV status
Concordance between IHC and PCR assays for detecting MCPyV was high (>90%; Supplementary Table S5). The AUC of the random forest classifier showed that gene expression signatures identified from RNA-seq profiles reliably predicted MCPyV status when either IHC- or PCR-determined MCPyV status was used as a classification label (Supplementary Table S6).
In both cohorts, responses were observed regardless of MCPyV status, although numerically higher ORR were observed in patients with MCPyV− versus MCPyV+ tumors. In patients with MCPyV+ versus MCPyV− tumors, ORR were 28.3% (13/46 patients) versus 35.5% (11/31 patients) in the 2L+ cohort and 38.1% (24/63 patients) versus 50.0% (18/36 patients) in the 1L cohort, respectively. The best overall response per MCPyV status in the 2L+ and 1L cohorts is shown in Supplementary Table S7. DNA damage repair–related pathways were enriched only in samples classified as MCPyV+ by PCR.
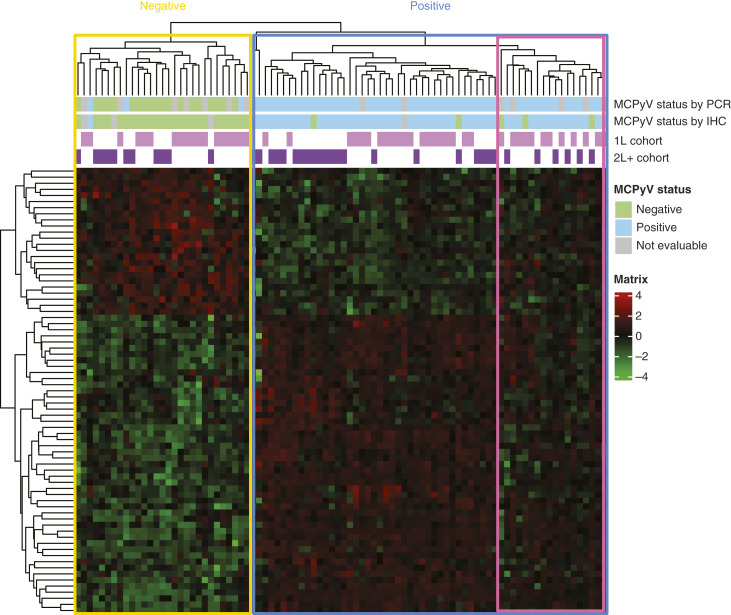
Patient samples were hierarchically clustered using 73 genes that were common in four gene expression signatures learned from classification experiments with a combination of MCPyV IHC and PCR labels from both cohorts. This novel 73-gene signature coherently clustered the samples according to MCPyV status (Fig. 2). As evident from a small subcluster with missing/discordant PCR and IHC labels, PCR classification of MCPyV status was more concordant with the expression cluster than IHC classification; MCPyV status was defined using IHC classification for all subsequent analyses.
Figure 2.
Clustering based on the 73-gene signature differentiates MCPyV status. The pink border highlights a cluster with discordant profiles compared with the rest of the MCPyV+ subgroup.
Immune cell profile by MCPyV status
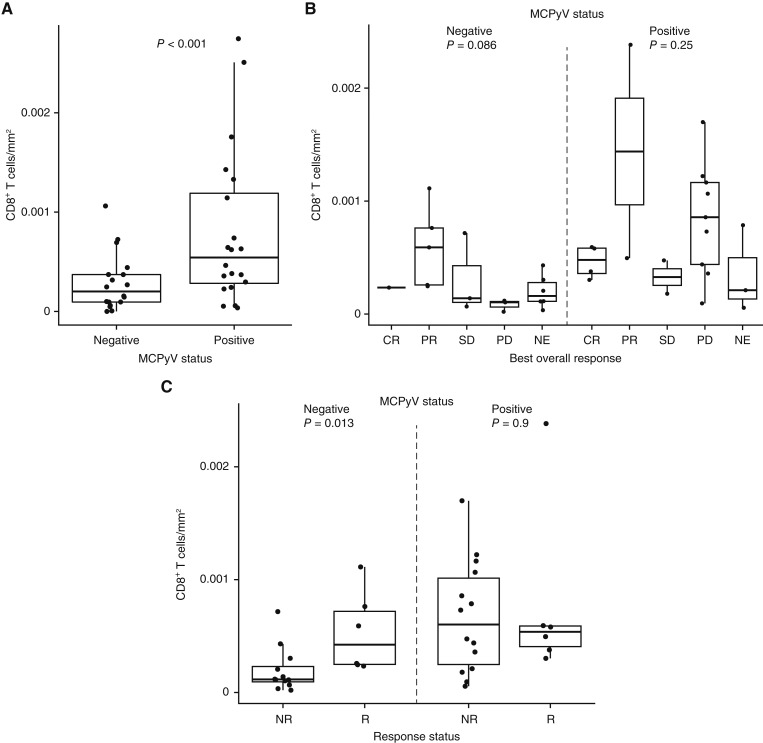
Higher CD8+ T-cell infiltration by digital pathology analysis was observed in MCPyV+ versus MCPyV− samples from the 2L+ cohort (Fig. 3A). In MCPyV− samples, a potential association was observed between higher CD8+ T-cell density and response (Fig. 3B). This potential association was also observed via RNA-seq analysis of MCPyV− samples from both 2L+ and 1L cohorts (Supplementary Fig. S6A and S6B). Progressive disease was observed in some patients despite high levels of CD8+ cells observed via digital pathology, suggestive of T-cell dysfunction. Responses in patients with MCPyV+ tumors were more common in patients with lower ICOS expression (P = 0.033; Supplementary Fig. S6C). Individual patients with MCPyV+ tumors seemed more likely to have a CD8+ T-cell density at or above the median (Supplementary Fig. S7). Immune content deconvolution based on RNA-seq profiles using different methods is shown in Supplementary Figs. S7 and S8. In both cohorts, patients with MCPyV+ versus MCPyV− tumors had greater infiltration of M2 macrophages (Supplementary Figs. S7A, S7B, S9A, and S10), and this increased infiltration was observed in conjunction with a higher expression of PD-L1 (Supplementary Figs. S9B and S10). No association between M2 macrophage infiltration and level of response was observed (Supplementary Fig. S10).
Figure 3.
Digital pathology analysis of CD8+ T-cell density at the invasive margin in samples from the 2L+ cohort according to (A) MCPyV status and (B and C) response and MCPyV status. CR, complete response; NE, not evaluable; NR, nonresponder; PD, progressive disease; PR, partial response; R, responder; SD, stable disease.
Comparison of biomarkers pre- and postchemotherapy
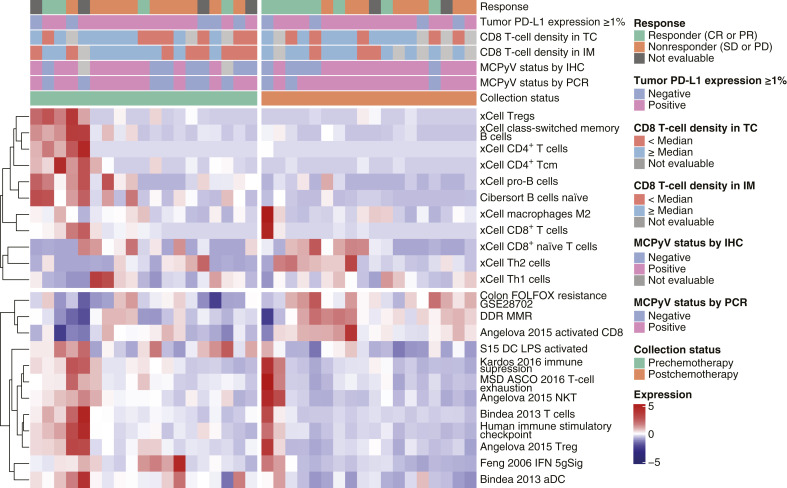
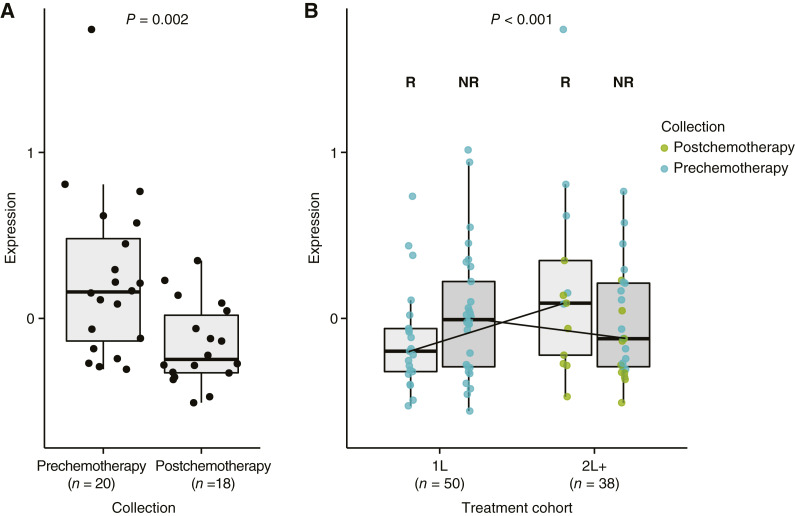
In the 2L+ cohort, postchemotherapy samples (vs. prechemotherapy samples) had enriched gene expression signatures for resistance to chemotherapy (including FOLFOX), DNA replication and repair, and cell-cycle regulation (including MYC and E2F targets), as shown in Fig. 4. Furthermore, inflammatory response signatures, such as IFNγ, were depleted. Both innate and adaptive immune responses seemed to be downregulated in postchemotherapy samples, with reduced levels of naïve B cells, subsets of T cells, and activated dendritic cells (DC). Higher levels of M2 macrophages were observed in postchemotherapy samples, suggesting an immunosuppressive environment; however, higher levels of activated CD8+ T cells, lower immune suppression, lower T-cell exhaustion, and the presence of regulatory T cells were also observed. Postchemotherapy samples also had lower expression of an activated DC signature (S15 DC LPS activated) compared with prechemotherapy samples (Figs. 4 and 5A). An ANOVA F test was used to assess the interaction between treatment line and response to avelumab, adjusting for other covariates. Using this test, the activated DC signature was found to be differentially present between patients with and without a response to avelumab (P < 0.001; Fig. 5B).
Figure 4.
Comparison of samples collected prechemotherapy (left) and postchemotherapy (right) from the 2L+ cohort and association with PD-L1 status, MCPyV status, CD8+ T-cell density, and immune cell infiltration. CR, complete response; DDR, DNA damage response; IM, invasive margin; LPS, lipopolysaccharide; MMR, mismatch repair; PD, progressive disease; PR, partial response; SD, stable disease; TC, tumor core; Treg, regulatory T cell.
Figure 5.
Levels of a signature for activated DC in (A) pre- and postchemotherapy samples from the 2L+ cohort and (B) responders and nonresponders in the 2L+ and 1L cohorts; interaction P values are shown. NR, nonresponder; R, responder.
Analysis of tumor microenvironment subtypes
In both the 2L+ and 1L cohorts, Bagaev signatures (24) were used to define tumor microenvironment (TME) subtypes: immune-enriched nonfibrotic (IE), immune-enriched fibrotic, and fibrotic; the desert (immune-depleted) subtype was not observed (Supplementary Fig. S11A and S11B). In tumors classified as having IE subtype, expression of gene signatures associated with angiogenesis and fibroblast signatures was generally lower, and expression of gene signatures associated with antitumor immunity was generally higher. To assess the potential association between TME and survival, the IE and immune-enriched fibrotic subgroups (immune-enriched tumors) were merged and compared with the fibrotic subgroup (fibrotic). OS and PFS observed in patients in the 1L cohort with immune-enriched tumors were longer than those in patients with fibrotic tumors (Supplementary Fig. S11C). In contrast, no differences were observed in the 2L+ cohort. Responses to avelumab were observed in patients with both immune-enriched and fibrotic tumors.
Immunologic marker expression in tumors and immune cells
To further explore the correlation between marker expression in tumors and M2 macrophages in MCPyV+ tumors and the role of different cell types in response to immune checkpoint inhibitor treatment, a comprehensive analysis of the MCC single-cell RNA-seq dataset from Paulson and colleagues (19) was performed. This dataset comprises single-cell RNA-seq profiles of tumors from two patients with MCPyV+ MCC. In MCC tumor cells, <1% had PD-L1 expression, whereas M2 macrophages and antigen-presenting cells had higher frequencies of PD-L1 expression (Supplementary Table S8). In addition, a potential correlation between PD-L1 and CD163 expression in tumor-associated cells was observed (Supplementary Fig. S12), suggesting expression of PD-L1 in tumor-associated macrophages.
Discussion
To our knowledge, this is the largest genomic study of patients with mMCC that included both WES and paired RNA-seq profiles to date. Using samples obtained from both cohorts of the JAVELIN Merkel 200 trial, we confirmed and extended previous studies showing biological differences between MCPyV-driven and UV-driven MCC tumors, including the low and high TMB, respectively, and the lower frequency of mutations in cancer drivers in MCPyV+ tumors. We also observed immunologic differences between MCPyV-driven and UV-driven tumors, including greater infiltration of CD8+ T cells and M2 macrophages in MCPyV+ tumors, in addition to evidence of reduced immunologic activity in tumor samples obtained after chemotherapy. By analyzing samples obtained from both cohorts of the trial, several hypotheses generated using samples from one cohort (mostly the 2L+ cohort) could be validated using the other cohort.
Our findings suggest that the presence of MCPyV exerts control of the host genome and transcriptome, creating a unique gene expression signature and leading to an immunosuppressive TME. WES showed a low number of mutations and overall TMB in MCPyV+ tumors, consistent with previous findings (3, 25). Validated gene expression signatures with high predictive power for MCPyV status could be derived from RNA-seq profiles. Interestingly, two patients in the 2L+ cohort had tumors that were MCPyV+ by PCR and IHC but otherwise resembled UV-driven tumors based on mutational profiles and gene expression signature, suggesting that the presence of the virus could be a passenger event. Thus, the gene expression signature may provide an alternative method to determine MCPyV status. Additional methods for classifying MCPyV status include multimodal analysis, tumor-associated mutation detection, and methods to assess viral DNA integration (18, 26, 27).
CD8+ T-cell density measurements derived using digital pathology, which provided increased resolution for immune cell distribution, suggested higher CD8+ T-cell density at the invasive margin in patients with MCPyV+ tumors, as seen in previous studies (5, 28); however, no correlation with response was seen, consistent with studies with another immune checkpoint inhibitor (11). This observation suggests that T cells, although increased in number, may be exhausted in these patients. Higher CD8+ T-cell density was also observed in patients whose tumors had a low mutation rate. Immune content deconvolution from RNA-seq profiles suggested increased infiltration of M2 macrophages, which are anti-inflammatory and immunosuppressive (29), in MCPyV+ tumors, and the M2 macrophage score correlated strongly with PD-L1 expression. Because MCC tumors assessed in this study, regardless of viral status, expressed low levels of PD-L1, the majority of PD-L1 is expected to have been expressed by cells in the microenvironment, including M2 macrophages. Although this could not be assessed in the current study, this suggestion is consistent with previous studies (30). In these patients, the antitumor activity of avelumab may be due to inhibition of PD-L1 expressed in the microenvironment.
Consistent with previous studies, tumorigenesis in patients with MCPyV− tumors seemed to be due to genomic changes based on higher mutation rates, higher TMB, and higher levels of signature 7 (UV signature; CC>TT, C>T mutations) observed in these tumors. Numerically higher ORR were observed in patients with MCPyV− versus MCPyV+ tumors, but this difference was not statistically significant. Additionally, we found that higher CD8+ T-cell density might be associated with response to avelumab in patients with MCPyV− tumors, although CD8+ T-cell densities were generally lower in MCPyV− versus MCPyV+ tumors.
In samples from both 1L and 2L+ cohorts, several genes were commonly mutated, suggesting their functional importance in MCC tumorigenesis. Commonly mutated genes included the tumor suppressors TP53 and RB1, as previously reported, in MCPyV+ and more frequently in MCPyV− tumors; dysregulation of these pathways is thought to contribute to MCC tumorigenesis (25, 31). Another commonly mutated gene was CSMD1, which is the target for human papillomavirus integrations and chromosome rearrangements in oropharyngeal squamous cell carcinoma (32). In patients with MCPyV− tumors in the 1L cohort, mutations in six genes seemed to be associated with a lack of response, of which RB1 had the strongest association (OR, 10.373), suggesting a potential role for RB1 in resistance to immunotherapy. Patients with mutations in CTNND2 seemed to have shorter survival, particularly in patients with MCPyV− tumors, and tumors with CTNND2 mutations had lower IFNγ expression. CTNND2 encodes δ-catenin, which is involved in adhesive junction functioning, protein translocation, and WNT signaling; CTNND2 mutations have previously been associated with tumorigenesis in various cancers (33). Interaction between the small tumor antigen of MCPyV and δ-catenin is thought to increase the transcription of genes involved in transformation and tumorigenesis through the binding of the transcription repressor Kaiso (34). Mutations in GRIN2A, which encodes the GluN2A protein (a regulatory subunit of the glutamate-gated N-methyl-D-aspartate receptor), also seemed to be associated with worse outcomes. In melanoma, GRIN2A mutations are common and have been correlated with shorter survival (35, 36), suggesting a more general role for GRIN2A in outcomes of patients with skin tumors. Overall, the association of these mutations with lack of response highlights their potential as biomarkers. However, further work is needed to assess the functional significance of the mutations.
Previous analyses from the JAVELIN Merkel 200 trial indicated that gene sets linked to the inflammatory response, such as IFNγ, were more active in patients who responded to avelumab compared to those who did not (8, 9). Analyses of pre- versus postchemotherapy samples reported here suggested that prior chemotherapy reduces IFNγ-based inflammation and downregulates both innate and adaptive immune responses. Postchemotherapy samples also seemed to have increased levels of M2 macrophages. Among patients who responded to avelumab, levels of activated DC were higher in the 2L+ cohort (patients with disease progression following prior chemotherapy) than in the 1L cohort (patients with no prior treatment for metastatic disease). This finding is consistent with recent work from Oh and colleagues (37), suggesting that DC, despite being outnumbered by macrophages, are a critical source of PD-L1 and that their ablation reduces tumor growth. This observation suggests that treatment-naïve patients may require lower antigen presentation to respond to avelumab than those previously treated with chemotherapy require. Therefore, the potential for benefits with avelumab treatment for mMCC may be greatest if it is administered as 1L therapy, consistent with prospective data (16, 38, 39). In analyses of the TME, patients with immune-enriched tumors in the 1L cohort, but not the 2L+ cohort, seemed to have longer survival than patients with fibrotic (non–immune-enriched) tumors, further supporting a hypothesis that prior chemotherapy may affect immune responses in MCC tumors.
In conclusion, the analyses reported here provide further insights into the biology of MCC, highlighting differences between UV-driven and MCPyV-driven tumors and showing evidence of an immunosuppressive microenvironment in MCPyV-driven tumors and immunosuppression in chemotherapy-treated tumors. We also identified potential associations between worse outcomes with avelumab and mutations in specific genes that have not been previously investigated in MCC. The limitations of the data reported, including the small cohort size (reflecting the rarity of MCC) and exploratory nature of the analyses, mean that caution is needed when interpreting these observations, and extrapolation to the clinical setting is not appropriate. However, these findings suggest that further studies to investigate novel treatment combinations that can mitigate immunosuppression in MCC tumors or target newly identified pathways may be warranted.
Supplementary Material
supplement
Acknowledgments
The authors thank the patients and their families, investigators, coinvestigators, and the study teams at each of the participating centers. This trial was sponsored by the healthcare business of Merck KGaA, Darmstadt, Germany (CrossRef Funder ID: 10.13039/100009945), and was previously conducted under an alliance between the healthcare business of Merck KGaA, Darmstadt, Germany, and Pfizer. Medical writing support was provided by Nucleus Global and was funded by the healthcare business of Merck KGaA, Darmstadt, Germany. The authors would like to thank Naveen Jami for performing analyses that helped identify the virus-specific gene signature and Krithika Krishnan for analyzing associations between somatic mutations and efficacy. P. Nghiem received support from NCI P01-CA225517.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Disclosures
S.P. D’Angelo reports other support from Adaptimmune, Amgen, GlaxoSmithKline, Immune Design, Immunocore, Incyte, Nektar, the healthcare business of Merck KGaA, Darmstadt, Germany, Bristol Myers Squibb, Deciphera, and Merck & Co. during the conduct of the study. C. Lebbé reports personal fees from Bristol Myers Squibb, Pierre Fabre, Sanofi, Novartis, Merck & Co. Amgen, the healthcare business of Merck KGaA, Roche, and Inflax outside the submitted work. A.S. Brohl reports grants from the healthcare business of Merck KGaA outside the submitted work. T. Mrowiec reports employment by and other support from the healthcare business of Merck KGaA during the conduct of the study. T. Leslie was an employee of EMD Serono at the time the study was conducted. S. Georges reports employment by EMD Serono during the conduct of the study. G. Güzel was an employee of the healthcare business of Merck KGaA at the time the study was conducted and reports other support and stock ownership outside the submitted work. P. Shah reports employment by and other support from EMD Serono during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
S.P. D’Angelo: Writing–original draft, writing–review and editing. C. Lebbé: Writing–original draft, writing–review and editing. P. Nghiem: Writing–original draft, writing–review and editing. A.S. Brohl: Writing–original draft, writing–review and editing. T. Mrowiec: Investigation, writing–original draft, writing–review and editing. T. Leslie: Investigation, writing–original draft, writing–review and editing. S. Georges: Investigation, writing–original draft, writing–review and editing. G. Güzel: Investigation, writing–original draft, writing–review and editing. P. Shah: Conceptualization, data curation, formal analysis, supervision, writing–original draft, project administration, writing–review and editing.
References
- 1. Becker JC, Stang A, DeCaprio JA, Cerroni L, Lebbé C, Veness M, et al. Merkel cell carcinoma. Nat Rev Dis Primers 2017;3:17077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu W, MacDonald M, You J. Merkel cell polyomavirus infection and Merkel cell carcinoma. Curr Opin Virol 2016;20:20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goh G, Walradt T, Markarov V, Blom A, Riaz N, Doumani R, et al. Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget 2016;7:3403–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wong SQ, Waldeck K, Vergara IA, Schröder J, Madore J, Wilmott JS, et al. UV-associated mutations underlie the etiology of MCV-negative Merkel cell carcinomas. Cancer Res 2015;75:5228–34. [DOI] [PubMed] [Google Scholar]
- 5. Harms PW, Patel RM, Verhaegen ME, Giordano TJ, Nash KT, Johnson CN, et al. Distinct gene expression profiles of viral- and nonviral-associated Merkel cell carcinoma revealed by transcriptome analysis. J Invest Dermatol 2013;133:936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harms KL, Zhao L, Johnson B, Wang X, Carskadon S, Palanisamy N, et al. Virus-positive Merkel cell carcinoma is an independent prognostic group with distinct predictive biomarkers. Clin Cancer Res 2021;27:2494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bavencio (avelumab) . Prescribing information. Rockland, MA, USA: EMD Serono; 2023. [Google Scholar]
- 8. D’Angelo SP, Bhatia S, Brohl AS, Hamid O, Mehnert JM, Terheyden P, et al. Avelumab in patients with previously treated metastatic Merkel cell carcinoma: long-term data and biomarker analyses from the single-arm phase 2 JAVELIN Merkel 200 trial. J Immunother Cancer 2020;8:e000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D’Angelo SP, Lebbé C, Mortier L, Brohl AS, Fazio N, Grob JJ, et al. First-line avelumab in a cohort of 116 patients with metastatic Merkel cell carcinoma (JAVELIN Merkel 200): primary and biomarker analyses of a phase II study. J Immunother Cancer 2021;9:e002646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giraldo NA, Nguyen P, Engle EL, Kaunitz GJ, Cottrell TR, Berry S, et al. Multidimensional, quantitative assessment of PD-1/PD-L1 expression in patients with Merkel cell carcinoma and association with response to pembrolizumab. J Immunother Cancer 2018;6:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nghiem PT, Bhatia S, Lipson EJ, Kudchadkar RR, Miller NJ, Annamalai L, et al. PD-1 blockade with pembrolizumab in advanced Merkel-cell carcinoma. N Engl J Med 2016;374:2542–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Topalian SL, Bhatia S, Amin A, Kudchadkar RR, Sharfman WH, Lebbé C, et al. Neoadjuvant nivolumab for patients with resectable Merkel cell carcinoma in the CheckMate 358 trial. J Clin Oncol 2020;38:2476–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ganesan S, Mehnert J. Biomarkers for response to immune checkpoint blockade. Annu Rev Cancer Biol 2020;4:331–51. [Google Scholar]
- 14. Kaufman HL, Russell J, Hamid O, Bhatia S, Terheyden P, D’Angelo SP, et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol 2016;17:1374–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nghiem P, Bhatia S, Lipson EJ, Sharfman WH, Kudchadkar RR, Brohl AS, et al. Durable tumor regression and overall survival in patients with advanced Merkel cell carcinoma receiving pembrolizumab as first-line therapy. J Clin Oncol 2019;37:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Knepper TC, Montesion M, Russell JS, Sokol ES, Frampton GM, Miller VA, et al. The genomic landscape of Merkel cell carcinoma and clinicogenomic biomarkers of response to immune checkpoint inhibitor therapy. Clin Cancer Res 2019;25:5961–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carter MD, Gaston D, Huang WY, Greer WL, Pasternak S, Ly TY, et al. Genetic profiles of different subsets of Merkel cell carcinoma show links between combined and pure MCPyV-negative tumors. Hum Pathol 2018;71:117–25. [DOI] [PubMed] [Google Scholar]
- 18. Starrett GJ, Thakuria M, Chen T, Marcelus C, Cheng J, Nomburg J, et al. Clinical and molecular characterization of virus-positive and virus-negative Merkel cell carcinoma. Genome Med 2020;12:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paulson KG, Voillet V, McAfee MS, Hunter DS, Wagener FD, Perdicchio M, et al. Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat Commun 2018;9:3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abbott CW, Boyle SM, Pyke RM, McDaniel LD, Levy E, Navarro FCP, et al. Prediction of immunotherapy response in melanoma through combined modeling of neoantigen burden and immune-related resistance mechanisms. Clin Cancer Res 2021;27:4265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patwardhan A, Harris J, Leng N, Bartha G, Church DM, Luo S, et al. Achieving high-sensitivity for clinical applications using augmented exome sequencing. Genome Med 2015;7:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. European Medicines Agency . Guideline on the investigation of subgroups in confirmatory clinical trials. [cited 2019 Jan 31]. London, UK: European Medicine Agency. Available from:https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-investigation-subgroups-confirmatory-clinical-trials_en.pdf. [Google Scholar]
- 23. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bagaev A, Kotlov N, Nomie K, Svekolkin V, Gafurov A, Isaeva O, et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 2021;39:845–65.e7. [DOI] [PubMed] [Google Scholar]
- 25. González-Vela MDC, Curiel-Olmo S, Derdak S, Beltran S, Santibañez M, Martínez N, et al. Shared oncogenic pathways implicated in both virus-positive and UV-induced Merkel cell carcinomas. J Invest Dermatol 2017;137:197–206. [DOI] [PubMed] [Google Scholar]
- 26. Moshiri AS, Doumani R, Yelistratova L, Blom A, Lachance K, Shinohara MM, et al. Polyomavirus-negative Merkel cell carcinoma: a more aggressive subtype based on analysis of 282 cases using multimodal tumor virus detection. J Invest Dermatol 2017;137:819–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haugg AM, Rennspiess D, zur Hausen A, Speel EJ, Cathomas G, Becker JC, et al. Fluorescence in situ hybridization and qPCR to detect Merkel cell polyomavirus physical status and load in Merkel cell carcinomas. Int J Cancer 2014;135:2804–15. [DOI] [PubMed] [Google Scholar]
- 28. Sihto H, Böhling T, Kavola H, Koljonen V, Salmi M, Jalkanen S, et al. Tumor infiltrating immune cells and outcome of Merkel cell carcinoma: a population-based study. Clin Cancer Res 2012;18:2872–81. [DOI] [PubMed] [Google Scholar]
- 29. Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel) 2014;6:1670–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dowlatshahi M, Huang V, Gehad AE, Jiang Y, Calarese A, Teague JE, et al. Tumor-specific T cells in human Merkel cell carcinomas: a possible role for Tregs and T-cell exhaustion in reducing T-cell responses. J Invest Dermatol 2013;133:1879–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cimino PJ, Robirds DH, Tripp SR, Pfeifer JD, Abel HJ, Duncavage EJ. Retinoblastoma gene mutations detected by whole exome sequencing of Merkel cell carcinoma. Mod Pathol 2014;27:1073–87. [DOI] [PubMed] [Google Scholar]
- 32. Gao G, Johnson SH, Vasmatzis G, Pauley CE, Tombers NM, Kasperbauer JL, et al. Common fragile sites (CFS) and extremely large CFS genes are targets for human papillomavirus integrations and chromosome rearrangements in oropharyngeal squamous cell carcinoma. Genes Chromosomes Cancer 2017;56:59–74. [DOI] [PubMed] [Google Scholar]
- 33. Lu Q, Aguilar BJ, Li M, Jiang Y, Chen YH. Genetic alterations of δ-catenin/NPRAP/Neurojungin (CTNND2): functional implications in complex human diseases. Hum Genet 2016;135:1107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vinueza JL. Elucidating mechanisms of transformation by Merkel cell polyomavirus tumor antigens. American Association for the Advancement of Science Annual Meeting. Philadelphia (PA); [cited 2022 Feb 17–20]. Abstract 30126. [Google Scholar]
- 35. D’mello SA, Flanagan J, Green T, Leung E, Askarian-Amiri M, Joseph W, et al. Evidence that GRIN2A mutations in melanoma correlate with decreased survival. Front Oncol 2014;3:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet 2011;43:442–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oh SA, Wu D-C, Cheung J, Navarro A, Xiong H, Cubas R, et al. PD-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer. Nat Cancer 2020;1:681–91. [DOI] [PubMed] [Google Scholar]
- 38. Topalian SL, Bhatia S, Hollebecque A, Awada A, Boer JPD, Kudchadkar RR, et al. Non-comparative, open-label, multiple cohort, phase 1/2 study to evaluate nivolumab (NIVO) in patients with virus-associated tumors (CheckMate 358): efficacy and safety in Merkel cell carcinoma (MCC). Cancer Res 2017;77(13 Suppl):CT074. Abstract CT074. [Google Scholar]
- 39. Weppler AM, Pattison A, Bhave P, De Ieso P, Raleigh J, Hatzimihalis A, et al. Clinical, FDG-PET and molecular markers of immune checkpoint inhibitor response in patients with metastatic Merkel cell carcinoma. J Immunother Cancer 2020;8:e000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplement
Data Availability Statement
The data generated in this study are available upon request from the sponsor. Any requests for data by qualified scientific and medical researchers for legitimate research purposes will be subjected to the healthcare business of Merck KGaA, Darmstadt, Germany's (CrossRef Funder ID: 10.13039/100009945) Data Sharing Policy. All requests should be submitted in writing to the data sharing portal of the healthcare business of Merck KGaA, Darmstadt, Germany's data sharing portal (https://www.emdgroup.com/en/research/our-approach-to-research-and-development/healthcare/clinical-trials/commitment-responsible-data-sharing.html). When the healthcare business of Merck KGaA, Darmstadt, Germany has a coresearch, codevelopment, or comarketing or copromotion agreement or when the product has been out-licensed, the responsibility for disclosure might be dependent on the agreement between parties. Under these circumstances, the healthcare business of Merck KGaA, Darmstadt, Germany will endeavor to gain agreement to share data in response to requests.