Extended Data Fig. 4 |. Spatial Analysis of Breast Cell Types with CODEX and smFISH.
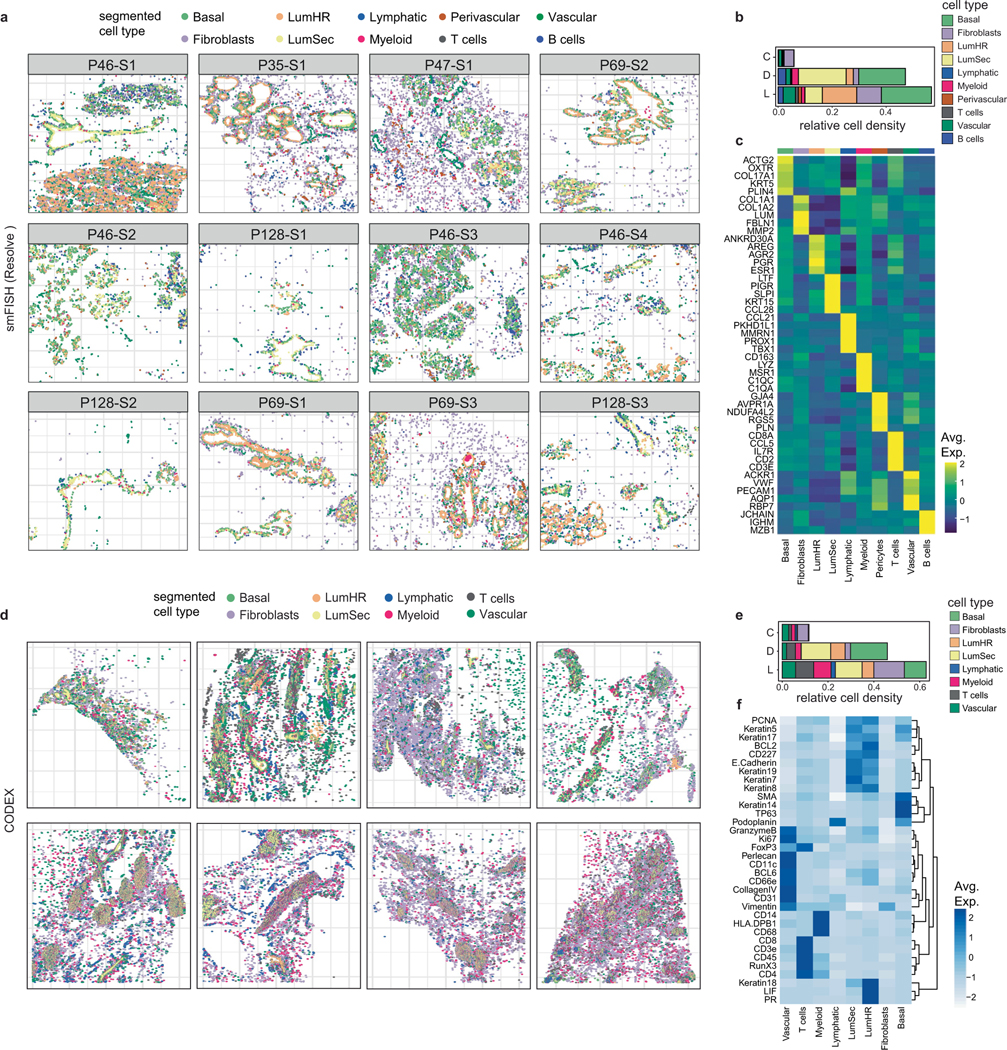
a, Cell segmentation results of smFISH (Resolve) data across 12 tissue samples profiled from 5 different women. Cells were annotated based on combinations of markers for each cell type as described in Supplementary Table 6. b, Densities of cell types across three topographic areas using 12 tissues profiled by smFISH (Resolve). c, Heatmap of the top 5 targeted maker genes for each cell type in the smFISH (Resolve) data from 12 combined tissue samples. d, Cell segmentation results of CODEX data from 8 different women. Cells were annotated based on combinations or single protein markers to identify different cell types. e, Densities of cell types across three topographic areas from 8 different women by CODEX. f, Heatmap showing protein levels for markers that were used to identify different cell types in the CODEX data. (D: ducts, L: lobules and C: connective regions).
